Quantitative PCR (qPCR) is a mature method for mRNA/DNA quantification and SNP screening, using fluorescent dyes or specifically labeled fluorescent nucleic acid probes to monitor the reaction process in real-time.
One critical decision in qPCR experiments is selecting the right chemistry: probes or dyes. The TaqMan probe vs SYBR dye debate is central to this decision. This guide breaks down their differences to help you choose reasonably.
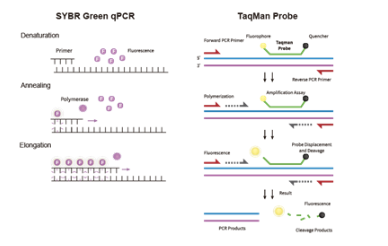
qPCR Chemistry Basics
In a standard PCR reaction, DNA is denatured, primers are annealed, and a polymerase extends the new strand.
qPCR (Quantitative PCR), or real-time PCR, builds on PCR by measuring DNA amplification in real time using fluorescent signals, allowing both detection and precise quantification of target sequences.
Detection Methods: Probes vs. Dyes
-
Dye-based Detection (SYBR Green): This method uses a non-specific fluorescent dye that binds all double-stranded DNA (dsDNA), emitting fluorescence proportional to total dsDNA content.
-
Probe-based Detection (TaqMan): This method utilizes a sequence-specific TaqMan probe that only generates a signal when it hybridizes to the exact target sequence between the two primers.
TaqMan Probes: How It Works
A TaqMan probe is a linear oligonucleotide designed to hybridize to an internal region of the target DNA, situated between the forward and reverse primers. It contains two critical components:
-
A Fluorophore Reporter: Usually located at the 5' end.
-
A Quencher: Usually located at the 3' end.
When the probe is intact, the physical proximity of the quencher to the reporter suppresses the fluorescent signal. During the PCR cycle’s extension phase, the Taq DNA polymerase (which possesses 5' to 3' exonuclease activity) encounters the annealed probe and cleaves it. This separation of the reporter from the quencher allows the reporter to emit fluorescence, proportional to the target DNA amplified.
1. Advantages of TaqMan
-
Exceptional Specificity: Because the signal is only generated if the qPCR probe binds to its specific target, the risk of false positives from non-specific amplification or primer dimers is virtually eliminated.
-
Multiplexing Capabilities: Unlike dye-based methods, you can use multiple probes labeled with different colored dyes in a single reaction tube. This allows for the simultaneous detection of several targets (e.g., a gene of interest and a housekeeping gene) in one well, saving both time and sample material.
-
No Melt Curve Needed: Since the chemistry is inherently specific, there is no requirement to perform a post-PCR dissociation curve to verify the product.
2. Limitations
The primary drawback of TaqMan qPCR is the complexity and cost. Each target requires a custom-synthesized probe. Furthermore, designing an effective probe involves strict parameters regarding melting temperature and sequence composition, making the initial setup more labor-intensive.
However, for researchers requiring accurate gene expression analysis or clinical diagnostics, TaqMan qPCR is often the gold standard.
SYBR Green I: How It Works
SYBR Green dye binds indiscriminately to all dsDNA in a reaction. These molecules emit very low fluorescence when they are free in solution. However, they have a high affinity for the minor groove of double-stranded DNA. Once they bind to dsDNA, their fluorescence increases several times. As the PCR reaction progresses and more double-stranded product is formed, the total fluorescence in the tube increases.
1. Advantages of SYBR Green
-
Cost-Effectiveness: You only need to purchase the standard primers and a master mix containing the dye.
-
Versatility: Since the dye binds to any dsDNA, a single master mix can be used for any target. This makes it an ideal tool for initial screening or when exploring many different genes in a non-model organism.
-
Simpler Design: Researchers only need to optimize two primers.
2. Limitations
The biggest challenge when comparing TaqMan vs SYBR is the issue of non-specific binding. SYBR Green will bind to any dsDNA, including:
-
Primer Dimers: Small artifacts formed by primers binding to each other.
-
Non-specific Amplicon: Unexpected DNA fragments resulting from poor primer design.
Because the dye cannot distinguish between the target and these artifacts, it can lead to overestimation of the target concentration or false-positive results. To mitigate this, a melt curve analysis must be performed after the PCR to ensure that only a single, specific product was amplified.
TaqMan vs SYBR: Key Differences
To help you decide which chemistry fits your project, the following table summarizes SYBR vs TaqMan setups.
| Feature | SYBR Green (Dye-Based) | TaqMan (Probe-Based) |
|---|---|---|
| Specificity | Low; detects all dsDNA | High; sequence-specific |
| Reproducibility | Moderate | High; very stable and consistent |
| Multiplexing | No; only one target per well | Yes; multiple targets per well (different dyes) |
| Cost | Low; economical for many targets | Higher; cost per target is more expensive |
| Design Complexity | Simple; requires only two primers | Complex; requires primers plus a specific probe |
| Validation Requirement | Requires melt curve analysis | No melt curve required |
TaqMan vs SYBR: How to Choose
Choosing between TaqMan vs SYBR chemistry depends on your specific experimental goals, your budget, and the level of precision required.
1. When to Choose SYBR Green
If you are in the early stages of a project, such as performing a large-scale screen of hundreds of potential gene candidates, SYBR Green is likely the better choice. It is also the preferred method for:
-
Method Development: Testing new primer pairs before committing to a probe design.
-
Low-Budget Research: When cost-per-reaction is a primary concern.
-
General Quantification: Applications like NGS library quantification, where the target sequence might vary.
2. When to Choose TaqMan
For clinical diagnostics, high-precision quantification, or high-stakes validation, a qPCR probe is the industry standard. Use TaqMan when you require:
-
Diagnostic Accuracy: In pathogen detection (like viral load monitoring), the high specificity of the TaqMan probe is non-negotiable to prevent false positives.
-
Allelic Discrimination: TaqMan is ideal for SNP genotyping, where a single nucleotide difference must be detected.
-
Limited Samples: If you have a very small amount of precious sample, multiplexing with TaqMan allows you to get more data points from a single reaction.
-
Multiplex detection: Multiple targets can be detected in a single reaction well because different targets can be labeled with probes carrying unique fluorescent dyes.
Diagnostic Solutions from Synbio Technologies
Whether your research demands the economical flexibility of SYBR Green or the rigorous precision of a TaqMan probe, the quality of your reagents is the ultimate predictor of success. Synbio Technologies provides a comprehensive suite of diagnostic probes and oligos to support your qPCR workflows.
Operating out of the ISO 9001 and ISO 13485 quality control management systems, Synbio Technologies specializes in:
-
Various diagnostic materials: FISH probes, qPCR probes, SNP probes, chemical modification groups, fluorescence, and quenchers.
-
Products in molecular hybridization and PCR: TaqMan/MGB qPCR probes, Molecular Beacons, Scorpions Probes, LNA Probes, Dual Quencher Probes, RAA/RPA Probes, etc.
You can customize diagnostic probes in various synthesis scales from nmol to umol at Synbio Technologies. They ensure high purity and low background noise. Each probe undergoes 100% LC-MS quality validation.
Feel free to contact us for any research needs!
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























