Synbio Technologies’ immune repertoire sequencing and bioinformatics analysis:
1. Library Construction
Synbio Technologies performed 5’ RACE reverse transcription of the extracted RNA and constructed the library.
2. Library Quality Control
The Qubit Fluorometer was used to perform preliminary quantification after library construction. Following dilution, insert size was determined based on the Agilent bioanalyzer. Once expected requirements were met, the effective concentration of the library (>2nM) was then accurately quantified by qPCR to ensure library quality.
3. Library Sequencing
Different libraries were pooled and sequenced by Illumina Miseq/HiSeq according to the effective concentration and the required amount of target data. Four fluorescent labeled dNTPs, DNA polymerase, and adaptors were added to the flow cell for the amplification. When each sequence cluster extended and generated the complementary chain, the corresponding fluorescent signals were released and captured after each new dNTP was added. The optical signals were then transformed into sequencing peaks by professional software to obtain the sequence information of the fragments.
4. Data Analysis
A quality assessment of the sequencing data was conducted.
|
|
|
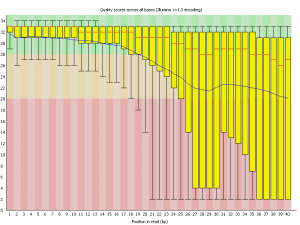
Per Base Sequence Quality
|
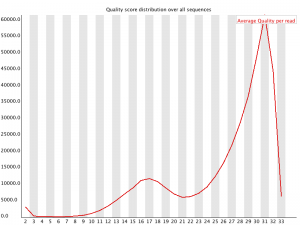
Per Sequence Quality Scores
|
|
|
|
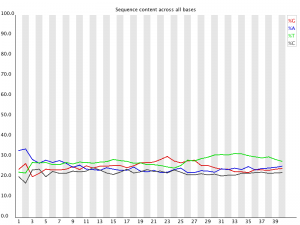
Per Base Sequence Content
|
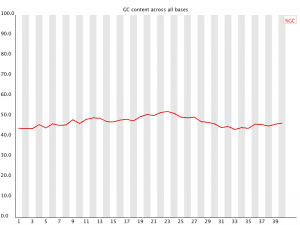
Per Base GC Content
|
|
|
|
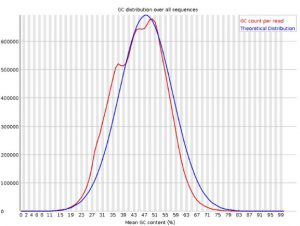
Per Sequence GC Content
|
Per Base N Content
|
|
|
|
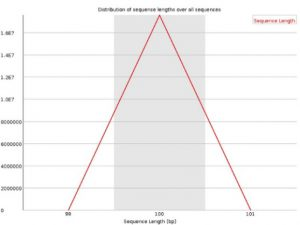
Sequence Length Distribution Reads
|
Duplicate Sequences
|
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products