The 2024 Nobel Prize in Chemistry was recently awarded to David Baker, Demis Hassabis, and John M. Jumper for their outstanding contributions to the fields of protein design and structure prediction. The DeepMind team’s development of AlphaFold3, with its powerful structural prediction unified framework, has opened new possibilities for drug development and has the potential to disrupt the current drug discovery model.
AI's role in protein research is rapidly evolving, with a primary focus on predicting protein structures and functions, understanding how proteins interact with other biomolecules, and advancing protein design and proteomics analysis. Notably, the combination of AI and protein design is leading the way in industrial applications, making significant strides in enhancing drug development and protein engineering processes. These advanced prediction techniques are increasingly integrated into existing workflows, streamlining research and innovation in the field. As AI continues to transform how we understand and manipulate proteins, its impact on biotechnology and medicine is poised to grow even further.
According to MedMarket Insights, the AI protein market size has reached 1,483M USD in 2023, and is expected to grow to USD 17.8B by 2031, at a CAGR of about 36.5% - primarily driven by the high adaptability of AI macromodels to life sciences.
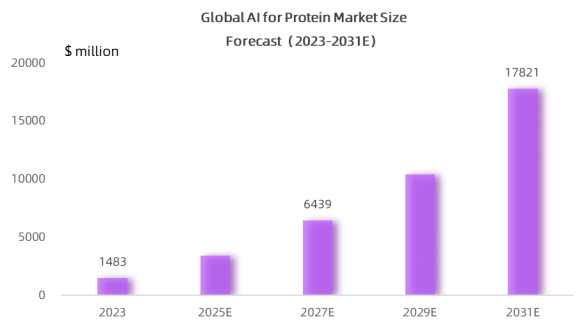
Source: MedMarket Insights, 2023
The U.S. is a global leader in AI protein research and applications, with about 58% of the market. Europe has 22%, Asia (mainly China and some Japanese companies) has about 12% market share, the UK and the Middle East have 4% and 2% respectively, and other regions have 2%.
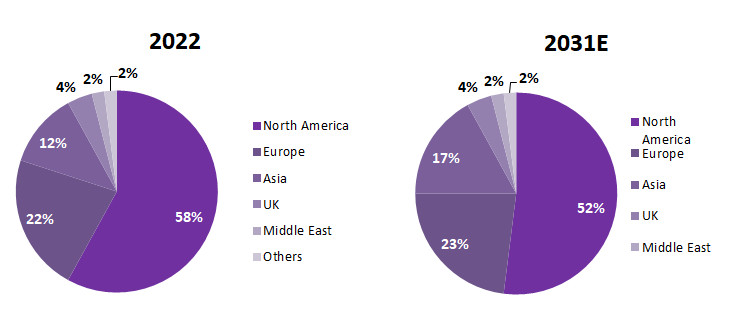
Global Prediction of AI for Protein Market Distribution
Source: MedMarket Insights, 2023
AI + Protein Application Scenarios
With the development of artificial intelligence (AI) technology represented by deep learning, it provides another way of thinking for protein research, bringing innovation in methods, processes and thinking. Especially with the breakthrough innovation represented by AlphaFold, AI has made significant progress in protein structure prediction, optimized design, and histological research. At the same time, these technologies have moved from the laboratory to practical applications, covering a wide range of fields such as protein design, drug discovery, synthetic biology, and more.
Protein Design
Artificial intelligence guided protein design can be divided into two major directions: protein optimization and protein design. Protein design from function to sequence requires only the identification of amino acid sequences that can perform that function. Protein optimization involves modifying a natural sequence (directed evolution) to enhance its specific function, such as affinity enhancement, catalytic activity improvement, and stability enhancement. Protein design, including sequence design from structure, sequence design from function and structure design from function, is an area in which Prof. David Baker and his team has been particularly successful.
Drug Discovery
AI protein prediction and protein design can greatly accelerate the process of new drug development. By predicting the structure and function of proteins alongside modeling the interaction process between proteins and other biomolecules, it is possible to accurately identify the binding site between a drug and its target. This provides very powerful support for drug development. In addition, end-to-end protein design technology based on artificial intelligence can realize the design of protein-based drugs with specific functions from scratch. At the same time, combining artificial intelligence with proteomics data can systematically analyze the association between proteins, diseases, and drugs in the human body - supplementing the target library and accelerating the process of drug discovery.
About 85% of human disease-related protein targets are difficult to become drugs, because some proteins are difficult to analyze and observe. Even when using traditional methods, such as electron microscopy and nuclear magnetic resonance, they can only observe their static structures. AI technology provides new tools and methods for drug discovery and development to address difficult-to-be-drugged targets, and is expected to change the traditional drug discovery model and increase the efficiency and success rate of drug discovery as a whole.
Synthetic Biology
In the area of synthetic biology, innovative solutions are provided for the agriculture, food, and pharmaceutical industries by precisely controlling and modifying the structure and function of proteins. This has huge application potential and economic value.
In the field of agriculture, AI can improve crop proteins, increase yield and quality, and reduce environmental pollution. 2021 NotCo Inc. launched Giuseppe, an AI platform that uses AI to design and predict the structure of plant-based proteins to mimic the nutrients and taste found in animal-based foods while providing optimization recommendations for the development of plant-based foods.
In the food sector, AI can help develop superior proteins that are more nutritious and less costly. Arzeda, a synthetic biology company, has achieved significant results in the field of enzyme modification, using their advanced enzyme modification platform to develop enzymes that efficiently convert sweeteners and reduce production costs. In addition, Arzeda has developed novel enzymes for BP to improve the efficiency of the oil extraction and production process.
How can AI-designed proteins be turned into functional proteins?
Turning AI-designed proteins into actual functional proteins involves several crucial steps.
1. Designing the Protein
• Functional Site Identification:The first step is to identify the functional site of the protein, which is responsible for its biochemical activity.
• Sequence Design: Using deep learning algorithms, researchers design an amino acid sequence that can fold into a three-dimensional structure containing the desired functional site. This step is particularly challenging as it requires creating a stable overall scaffold with a functional region (such as active sites or binding interfaces) and designing a sequence that folds into this structure.
2. Validation and Optimization
• Validation: Before proceeding to experiments, the designed protein is validated computationally. This includes checking the stability of the protein, ensuring that the functional site is correctly positioned, and predicting its interaction with other molecules.
• Optimization: Based on computational validation, the design may undergo several rounds of optimization to improve its stability, functionality, and expression in biological systems.
3. Synthesis and Expression
Gene Synthesis: Once the optimized protein sequence is obtained, the next step is to synthesize the corresponding gene. This can be done using DNA synthesis techniques, which are now highly automated and efficient.
Expression: The synthesized gene is then cloned into a suitable host cell (such as bacteria, yeast, or mammalian cells) for protein expression. The host cell is cultured under optimal conditions to promote high-level expression of the desired protein.
4. Purification and Characterization
Purification: After expression, the protein is purified from the host cell using various biochemical techniques, such as chromatography and electrophoresis. This step ensures that the protein is free from contaminants and impurities.
Characterization: The purified protein is then characterized using various biochemical and biophysical methods, such as spectroscopy, electrophoresis, and mass spectrometry. These methods provide information about the protein's structure, stability, and functional properties.
5. Functional Testing
Biological Activity Assays:The functional properties of the protein are tested using specific assays that measure its activity or binding affinity to target molecules. These assays can include enzymatic activity measurements, binding experiments, or cell-based assays.
Synbio Technologies | Bringing AI-designed Proteins to Life
Synbio Technologies combines the power of AI design and synthetic biology to provide comprehensive support from protein sequence optimization to expression to functional validation, facilitating downstream applied research on protein prediction, bringing AI-designed proteins to life.
Syno GS platform: AI-Driven Deep DNA Synthesis
Our Syno GS platform features a variety of intelligent biological analysis tools, including Syno Ab, NG Codon, Complexity Index (CI), and AI-TAT. You only need to provide the nucleotide or amino acid sequence you wish to synthesize, and we will deliver a 100% accurate gene sequence - cloned into your specified vector. Precise gene sequences can improve the accuracy and efficiency of protein expression.
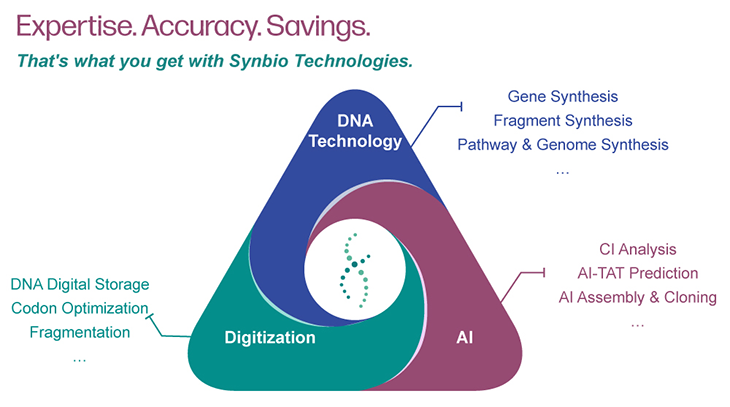
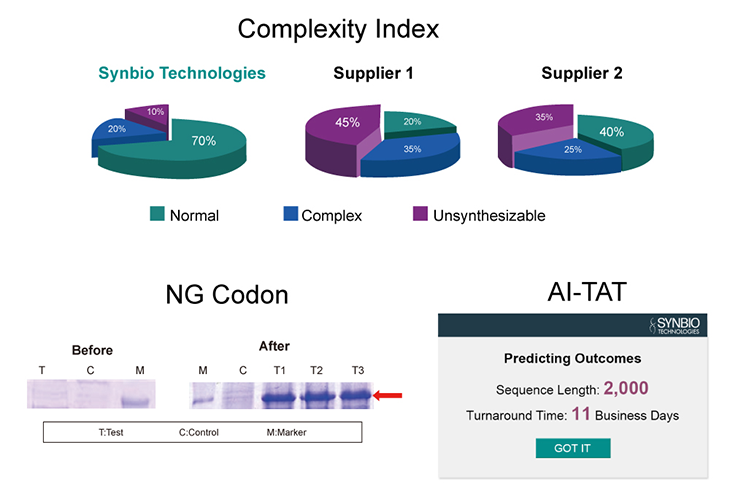
AI-Driven Deep DNA Synthesis
Syno Ab Platform: Enabling Antibody Discovery
AlphaFold excels at predicting protein structures but cannot simulate the changes in proteins interacting with other proteins or drug molecules. Synbio Technologies' Syno Ab antibody platform addresses this limitation by using the antigen structures predicted by AlphaFold as a starting point. Supported by antibody biological medicine biocomputing, it effectively simulates antigen-antibody docking, combining computational technology with experimental approaches to help researchers significantly reduce the overall costs of antibody development and shorten the development cycle.
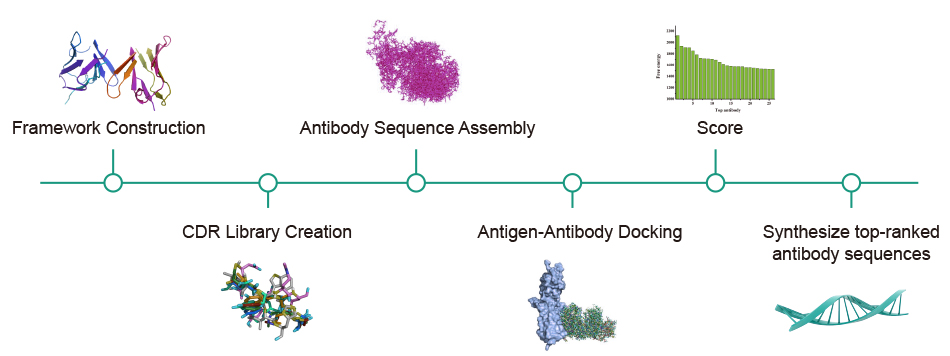
Service Process
Syno Protein Platform: From Sequence to Structure to Function
Synbio Technologies provides protein expression services for four expression systems: bacterial, yeast, insect, and mammalian. From milligrams to grams, our synthetic biology platforms and NG Codon optimization technology produce high-purity, active proteins to accelerate your research. Depending on the nature of the protein in question and the specific requirements of our customers, we carefully select from a range of expression vectors, expression hosts, and fusion tags to deliver optimal results. We proudly provide customized services to scientists around the world.
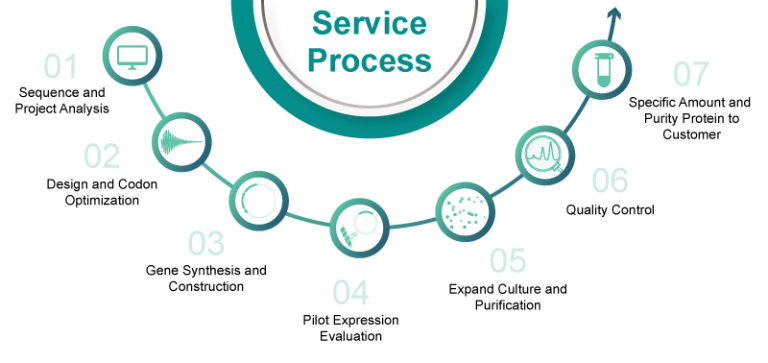
Protein Expression Service Process
References
1. ABRAMSON, Josh, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 2024, 1-3.
2. CHEN, Lingtao, et al. AI-Driven Deep Learning Techniques in Protein Structure Prediction. International journal of molecular sciences, 2024, 25.15: 8426.
3. GOEL, Divya; KUMAR, Ravi; KUMAR, Sudhir. AI-Assisted Methods for Protein Structure Prediction and Analysis. In: Microbial Data Intelligence and Computational Techniques for Sustainable Computing. Singapore: Springer Nature Singapore, 2024. p. 365-391.
4. Yoshida Hiroyuki,[Deep Learning and AlphaGo].[J] .Brain Nerve, 2019, 71: 681-694.
5. Niazi Sarfaraz K,Molecular Biosimilarity-An AI-Driven Paradigm Shift.[J] .Int J Mol Sci, 2022, 23.
6. Liu Z, Zhang Y, Nielsen J. Synthetic biology of yeast[J]. Biochemistry, 2019, 58(11): 1511-1520.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























