General Introduction of sgRNA Customized Design
sgRNA (single-guide RNA) is a key component in the CRISPR-Cas9 gene-editing system. It is a short, synthetic RNA sequence that directs the Cas9 enzyme to a specific DNA target for cutting. sgRNA can be designed for different CRISPR applications, such as gene knockout, base editing, and epigenetic modifications.
The sgRNA consists of two key regions:
-
Guide sequence (20 nucleotides) – Binds to the complementary DNA target via base pairing.
-
Scaffold sequence – Binds to the Cas9 enzyme, ensuring proper complex formation and function.
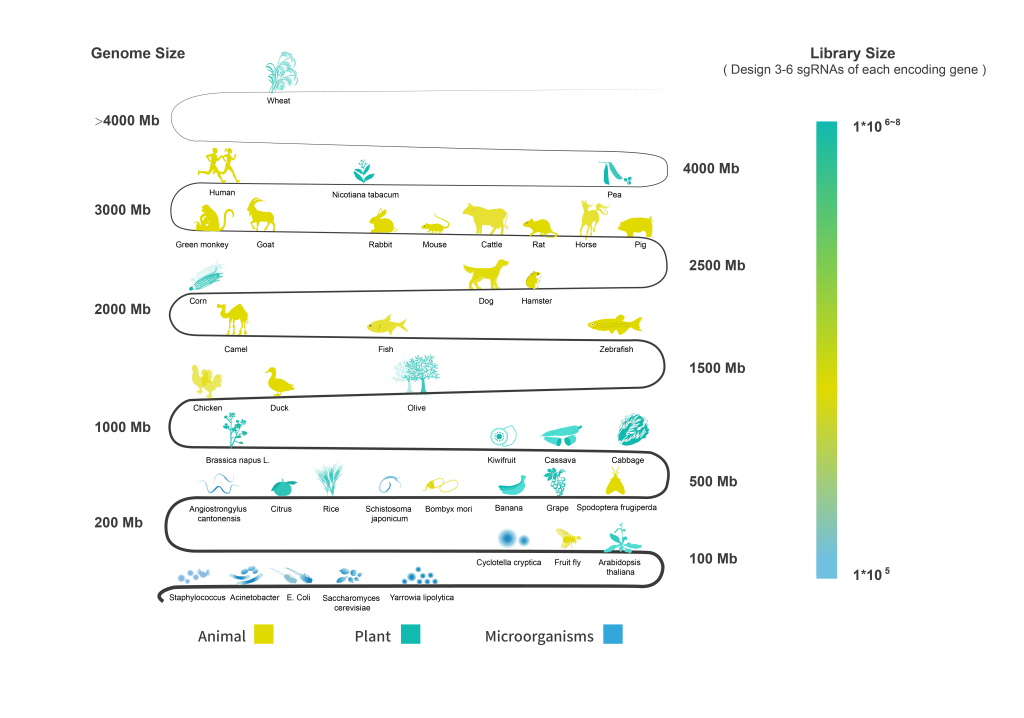
Synbio Technologies provides single sequence sgRNA design and whole-genome sgRNA library design, downstream verification, and stable cell line construction services. We offer a one-stop solution for CRISPR projects to achieve high genome editing efficiency.
Highlights
-
![]()
- Well-designed Sequences
-
![]()
- More Than 20 Designed Species
-
![]()
- Customized sgRNAs
-
![]()
- Single sgRNA, Multiple sgRNA and sgRNA Library Design Services
Service Details
|
Service Program
|
Service Details
|
TAT
|
|
sgRNA design
|
The gRNA sequences are designed for specific gene classifications, with 3-6 sgRNAs designed for each gene
|
1-2 weeks
|
The Difference Between gRNA and sgRNA View More
The main difference between gRNA (guide RNA) and sgRNA (single-guide RNA) lies in their structure and use in the CRISPR-Cas9 system:
-
gRNA (guide RNA) – This is a general term that refers to the RNA molecule that guides the Cas enzyme to a specific DNA target. In the natural CRISPR system (bacteria and archaea), the gRNA consists of two separate RNA molecules:crRNA and tracrRNA.
-
sgRNA (single-guide RNA) – This is a synthetic, engineered version of gRNA used in laboratory applications. It combines crRNA and tracrRNA into a single RNA molecule, simplifying the system and making it more efficient for gene editing.
FAQs
A unique primer binding site can be inserted into the screening library plasmid.
1. The GC content must be between 40% and 80% to ensure stable binding between the sgRNA and the target DNA.
2. Removal of polyA sites, which disrupt viral packaging if viruses are used for delivery.
3. Ensure that sgRNA designs have fewer secondary structures, such as hairpin structures and polymerase termination sequences; these structures can affect cloning efficiency and guide transcription.
How to Design Your sgRNA for CRISPR Genome Editing?
In gene-editing experiments, sgRNA is preferred because it is easier to design, synthesize, and use for targeted modifications. Designing an effective single-guide RNA (sgRNA) for CRISPR genome editing is crucial for achieving high specificity and efficiency. Here’s a step-by-step guide to designing sgRNA:
1. Select Target Gene and Region
Identify the gene or sequence you want to edit. Choose a target site near the region of interest while considering the availability of a PAM (Protospacer Adjacent Motif) sequence.
2. Use an Online sgRNA Design Tool
Use bioinformatics tools like Benchling or CHOPCHOP to find high-efficiency sgRNA candidates while minimizing off-target effects.
3. Check sgRNA Design Criteria
A good sgRNA should be 20 nucleotides long, have 40-60% GC content, avoid homopolymeric sequences, and target the non-template strand if applicable.
4. Check for Off-Target Effects
Use online tools to scan the entire genome for unintended binding sites. Prioritize sgRNAs with fewer or no off-targets (especially in coding regions). Modify mismatches near the 5’ end if needed to reduce off-target binding.
5. Synthesize sgRNA or CRISPR Vector Construction
Once selected, the sgRNA can be synthesized or cloned into a CRISPR vector for expression.
6. Validate sgRNA Efficiency In Vitro
Perform a T7E1 assay or Surveyor assay to check cleavage efficiency. Use Sanger sequencing or NGS to confirm desired mutations. Optimize by testing multiple sgRNAs if needed.
Get in Touch with Us
Related Services
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome KO Plasmid
Standard Genome KO Plasmid ProXpress
ProXpress Protein Products
Protein Products






























