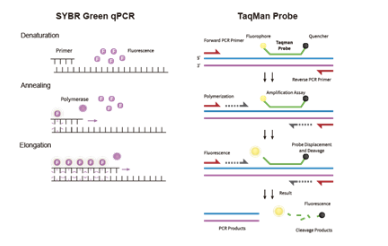
During the qPCR experiment using probes, the strength of the probe's fluorescence signal represents the degree of amplification and the quantity of a specific DNA sequence in the reaction system. Each time a DNA strand is amplified, a fluorescence molecule is formed, resulting in the accumulation of the fluorescence signal, which is perfectly synchronized with the formation of PCR products. Therefore, the fluorescence signal strength is directly proportional to the amount of double-stranded DNA in the reaction system. As the PCR reaction progresses, the target DNA sequence continues to be amplified, and the fluorescence signal increases accordingly.
What does the strength of the probe's fluorescence signal mean?
1. Quantitative Analysis
By real-time monitoring of the changes in fluorescence signal strength, a fluorescence amplification curve can be plotted. Based on the shape and characteristics of the curve, quantitative analysis of the target DNA in an unknown sample can be performed. For example, during the exponential growth phase, there is a linear relationship between the logarithmic value of PCR product quantity and the initial template amount, which is the optimal period for quantitative analysis. At this stage, the fluorescence signal strength can directly reflect the amount of the initial template.
2. Cycle Threshold (Ct) Value
The fluorescence signal strength is used to calculate the Ct value. The lower the Ct value, the higher the initial amount of target DNA or RNA. Conversely, a higher Ct value indicates a lower initial amount of target. By comparing the Ct value of unknown samples with standard curves generated from known quantities of the target, the initial concentration of the target in the sample can be calculated.
3. Reaction Efficiency and Specificity
The strength of the fluorescence signal can also reflect the efficiency of the PCR reaction. If the fluorescence signal is weak or increases slowly, it may indicate the presence of inhibitors in the reaction system or issues with primer design.
Additionally, because qPCR probes are highly specific, the fluorescence signal strength can also serve as an indicator of the reaction's specificity. If non-specific amplification leads to an increase in the fluorescence signal, it may indicate problems with the experimental design or procedure. Therefore, by real-time monitoring of the changes in fluorescence signal strength, the target DNA in an unknown sample can be quantitatively analyzed, and the efficiency and specificity of the PCR reaction can be assessed.
So what factors affect the fluorescence signal intensity of qpcr?
1. Probe Design and Sequence
The specific design of the probe, including its length, GC content, and sequence, can significantly affect the fluorescence signal. Probes with higher GC content generally bind more strongly to the target sequence, improving signal intensity.The position of the probe relative to the PCR amplicon is crucial. The probe should be positioned between the forward and reverse primers to ensure efficient hybridization and fluorescence emission.
2. Fluorophore and Quencher Choice
The type and properties of the fluorophore (the molecule that emits fluorescence) and the quencher (which absorbs fluorescence emitted by the fluorophore) play a critical role in signal intensity. For effective quenching, the distance between the fluorophore and quencher should be optimal.The choice of fluorophore should match the detection capabilities of the qPCR machine, considering factors like emission wavelength and sensitivity.
3. Probe Concentration
Too high or too low probe concentrations can affect the efficiency of fluorescence emission. High probe concentrations may cause self-quenching or non-specific binding, while too low concentrations may result in weak signals and reduced sensitivity.
4. Amplification Efficiency
The efficiency of the PCR amplification affects the accumulation of target DNA during the reaction. Poor amplification efficiency can lead to low or inconsistent fluorescence signals. Optimal primer and probe concentrations, as well as a suitable reaction mixture, are essential for high amplification efficiency.
5. Reaction Conditions
Temperature and cycling conditions impact the efficiency of probe hybridization and PCR amplification. Suboptimal temperatures may reduce probe binding efficiency, while inappropriate cycling times can affect the fluorescence signal.
6. DNA Template Quality and Quantity
The amount and quality of the DNA template used in the qPCR reaction will influence the fluorescence signal. Degraded or insufficient template amounts can lead to weak or absent signals. The presence of inhibitors (such as salts, detergents, or residual solvents) in the template DNA can interfere with the probe's ability to bind to the target and generate a signal.
7. Background Fluorescence and Instrument Sensitivity
Background fluorescence or instrument noise can mask or interfere with the fluorescence signal from the probe. Ensuring that the qPCR machine is properly calibrated and that the reagents are of high quality can minimize this issue. The sensitivity of the qPCR instrument, including its ability to distinguish low levels of fluorescence, is crucial for accurate quantification.
8. Efficiency of Probe Hybridization
Probe hybridization efficiency can be affected by sequence mismatches, secondary structures in the target DNA, and the melting temperature (Tm) of the probe-target complex. If hybridization is inefficient, the fluorescence signal will be weaker.
9. Competition with Primer Binding
The probe must bind specifically to its target sequence, but if primers are present at very high concentrations, they may outcompete the probe for binding sites, leading to reduced probe-target hybridization and weaker fluorescence signals.
The above are mainly the factors affecting the fluorescence signal of qPCR. In order to obtain accurate and reliable qPCR results, it is necessary to consider these factors comprehensively and take corresponding measures to control and optimize them.
Synbio Technologies | Diagnosis Probe Services
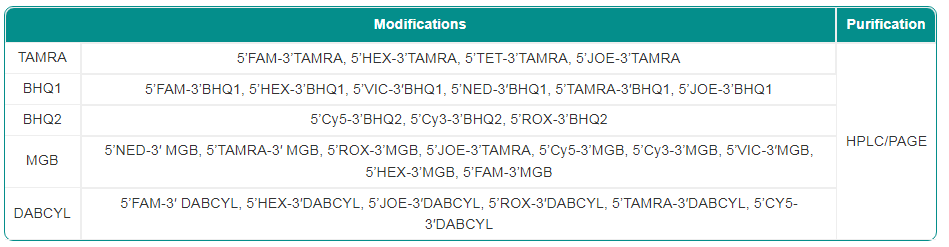
Synbio Technologies provides high quality nucleic acid probes with batch-to-batch consistency for qPCR/SNP detection. Our probes can also be labeled at the 5′ and 3′ ends of a designated position according to the specific needs of our customers. Depending on your project, we can modify these probes in order to improve the efficiency, accuracy, and diversity of qPCR assays.
Service Detail
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























