At Synbio Technologies, we often meet researchers who struggle with qPCR Probe Selection because long-standing assumptions continue to influence how assays are designed. Many users rely on familiar templates without reconsidering how sequence structure, target behavior, or instrument settings affect performance. As we support scientists working with qpcr probe design for diverse applications, we see how these misunderstandings can slow progress. Since Real-time Quantitative PCR relies on precise signal interpretation, clarifying these misconceptions can help users refine their experiments with greater confidence.
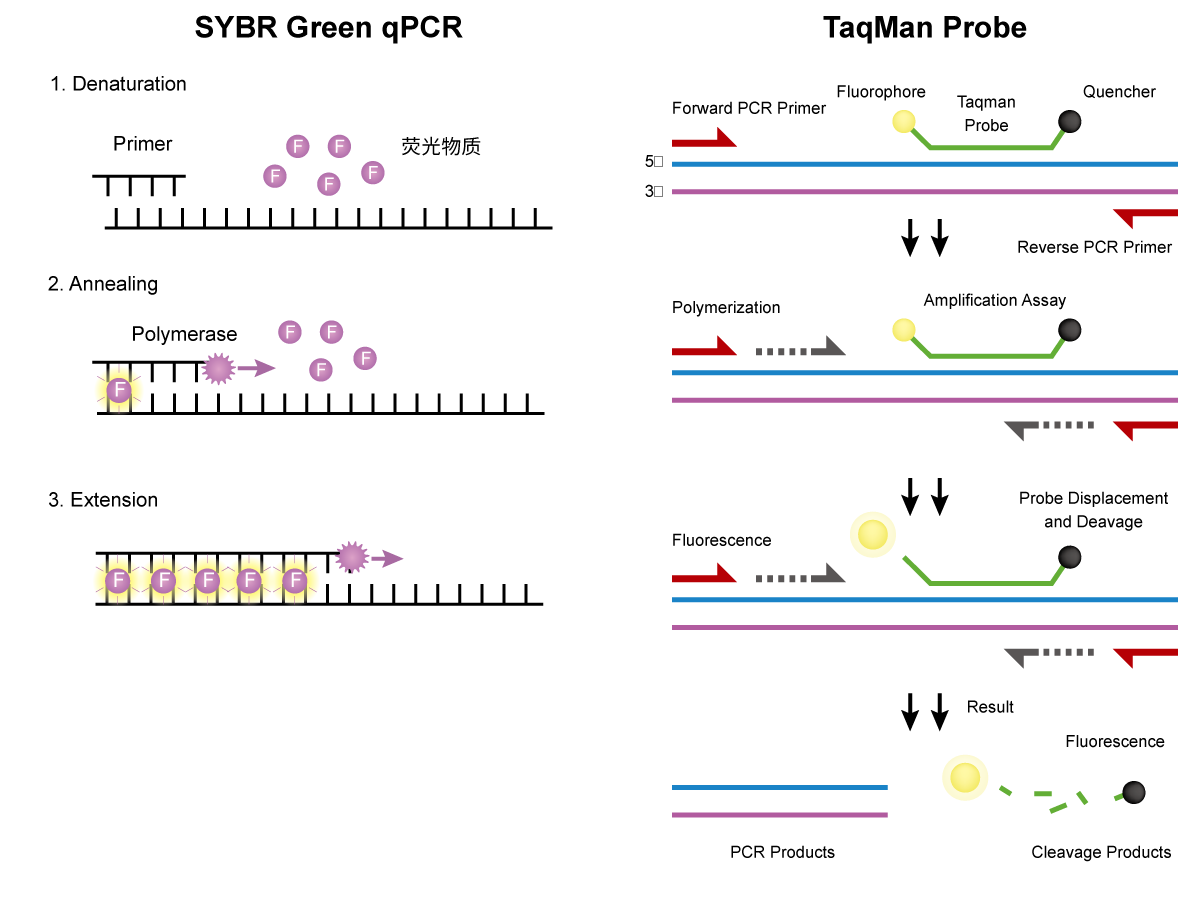
A Universal Probe Design Fits Every Target
One frequent misunderstanding is the belief that a single design approach works equally well across any sequence. When we review projects that involve qPCR Probe Selection, we often find that subtle factors—such as GC balance, secondary structure, and target accessibility—shape how a qpcr probe interacts with the template. Because Real-time Quantitative PCR depends on stable hybridization, a design that works for one gene may not behave consistently with another. In our own workflow, we evaluate probe regions with attention to melting characteristics and contextual sequence features rather than relying on one fixed formula. This helps establish predictable performance when researchers shift between pathogen detection, gene expression studies, or stability assessments.
Shorter Probes Always Produce Cleaner Signals
Another misconception is that shorter probes automatically generate clearer fluorescence. When users consult us about qPCR Probe Selection, they sometimes assume that fewer nucleotides translate into less background. However, the interaction between length, specificity, and quenching efficiency is more nuanced. A qpcr probe that is too short may bind non-specifically, while one that is moderately longer can improve discrimination between similar targets. Since Real-time Quantitative PCR requires balanced sensitivity, we analyze each target region to identify a length that supports selective binding without compromising overall dynamics. This approach aligns with our design recommendations described in our internal guidelines for selecting suitable probes for different assay types.
Probes Do Not Need Validation if Primers Work Well
Some users believe that successful primers guarantee an equally successful probe. In our experience, this assumption disrupts qPCR Probe Selection by overlooking how the probe contributes to overall assay behavior. A qpcr probe must maintain its own structural integrity, remain free from unintended interactions, and function predictably under cycling conditions. Even when primers meet basic criteria, Real-time Quantitative PCR performance may decline if the probe introduces fluorescence variability. To address this, we validate probe candidates through sequence screening, structural review, and experimental checks. Our company incorporates these steps into our design-to-use workflow so researchers receive components that support consistent amplification curves.
Conclusion: Correcting Misunderstandings for Better Assay Outcomes
Clearing up these misconceptions helps researchers refine assay planning and reduce unnecessary troubleshooting. When we revisit qPCR Probe Selection with a focus on sequence context, structural behavior, and functional validation, the resulting designs support more dependable experimentation. Our evaluation process clarifies how each qpcr probe influences specificity and signal quality, contributing to more stable Real-time Quantitative PCR results. At Synbio Technologies, we continue to assist users in selecting and applying probe designs that align with real scientific requirements rather than common but misleading assumptions.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























