Unlocking the Genome with High-Throughput shRNA Screening
In modern functional genomics research, the ability to systematically explore the genome is crucial for unraveling disease mechanisms and identifying new therapeutic strategies. One of the most effective technologies enabling this is the short hairpin RNA (shRNA) library. By leveraging the natural process of RNA interference (RNAi), shRNA libraries allow scientists to silence thousands of genes in parallel, offering a high-throughput approach to decode gene function, uncover novel drug targets, and accelerate cancer research.
What Is shRNA Library?
While individual shRNA experiments can reveal the role of a single gene, biology rarely works in isolation. Complex processes such as drug resistance, cell differentiation, tumorigenesis, and viral infection are driven by interconnected gene networks. To fully understand these networks, researchers need a technology that can simultaneously and systematically perturb thousands of genes—this is where shRNA library screening becomes essential.
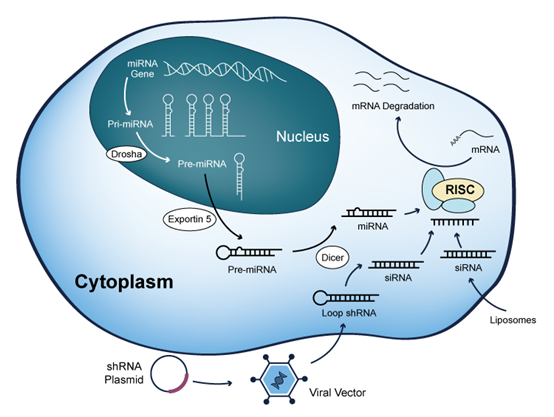
An shRNA library is a comprehensive collection of tens of thousands of unique shRNA sequences, delivered via lentiviral vectors. Each sequence targets a specific gene transcript, enabling genome-wide knockdown studies. With robust viral packaging and uniform coverage, these libraries provide a scalable solution for genome-wide RNAi screening.
How Do shRNA Libraries Work?
One of the most powerful applications of shRNA libraries is large-scale loss-of-function screening. Scientists can use the library to answer the question at the cellular level: “Which genes, when silenced, will result in a specific phenotype of interest from the tens of thousands of genes?”
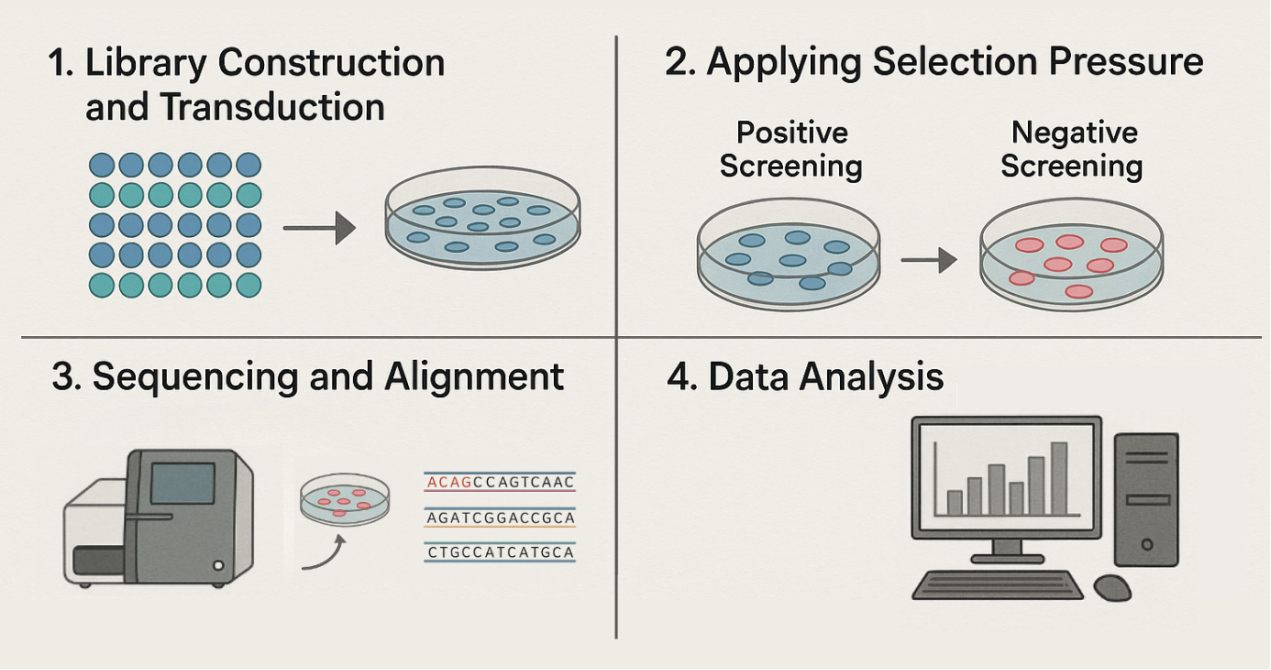
- 1. Library Construction and Transduction:
A viral library containing tens of thousands of unique shRNA sequences is introduced into a large population of cells, ensuring that each cell receives only 1-2 shRNAs randomly, thus forming a cell pool in which every gene is silenced to varying degrees.
- 2. Applying Selection Pressure:
Specific selection conditions are then applied to this cell population.
- Positive Screening: For instance, adding an anticancer drug to the culture. Cells that survive (due to the silencing of a specific gene leading to drug resistance) or proliferate quickly will become enriched.
- Negative Screening: For example, identifying genes critical for cell survival. Cells that die or disappear due to the silencing of essential genes will be eliminated. These cells are abundant in the population before screening but will be significantly depleted after screening.
- 3. Sequencing and Alignment:
After the screening process, genomic DNA from the surviving cell population is extracted, and high-throughput sequencing technology is used to analyze the shRNA sequences and their abundance.
- 4. Data Analysis:
By comparing with the original library before screening, researchers can identify shRNAs that are significantly enriched or depleted in the surviving cells. The genes targeted by these shRNAs are closely associated with the phenotype under study, such as drug resistance or cell survival.
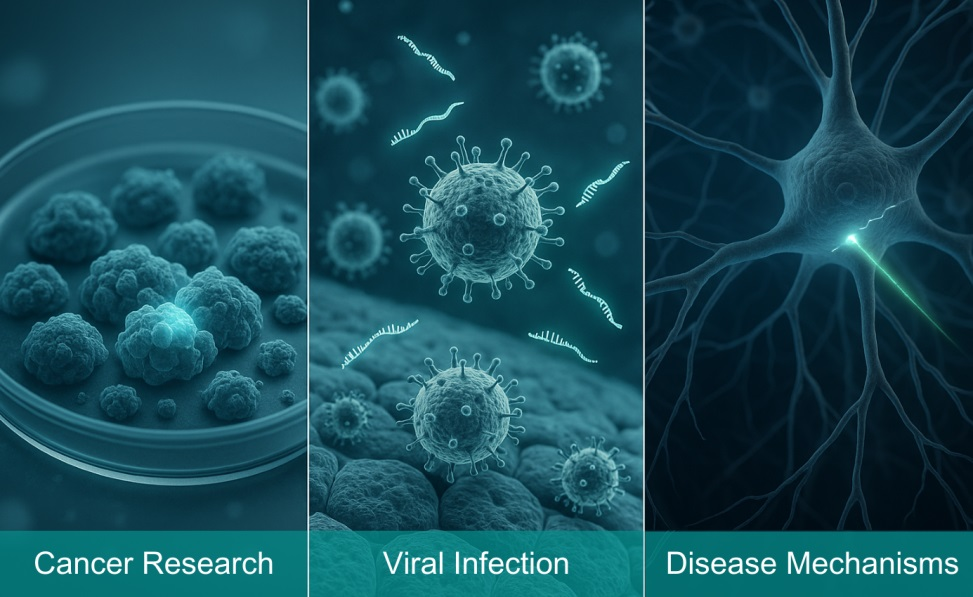
Applications of shRNA Libraries
Discovery of New Drug Targets or Disease-Related Genes
- Cancer Research:
Screening for genes whose silencing makes cancer cells more sensitive to chemotherapy drugs. These genes may themselves be excellent drug targets (if inhibiting the protein enhances drug efficacy).
- Viral Infections:
Screening for genes in host cells that are essential for viral replication, providing new insights for the development of antiviral drugs.
- Disease Mechanisms:
Screening for genes that play a key role in specific disease models (such as neurodegenerative or metabolic diseases), uncovering new disease mechanisms.
Synthetic Lethality Screening
This is an important concept in cancer therapy: two genes may individually mutate without causing cell death, but when both are mutated simultaneously, the cell dies. shRNA libraries can be used to identify genes that form a synthetic lethality relationship with known oncogenes (such as KRAS, MYC). This means that for cancers carrying specific oncogene mutations, we can design drugs to target another gene that is synthetically lethal with it, thereby specifically killing cancer cells without harming normal cells.
Validating Gene Function Networks
By analyzing gene sets that are enriched or depleted in screening results, bioinformatics analyses (such as pathway enrichment analysis) can be performed to identify the biological pathways these genes mainly participate in (e.g., MAPK signaling, apoptosis pathways, etc.), providing a system-level understanding of the gene functional networks that regulate a specific phenotype.
Key Advantages of shRNA Libraries
- High-Throughput Genomic Screening – Test tens of thousands of genes in a single experiment.
- Unbiased Discovery – Explore the whole genome without prior assumptions.
- Stable Gene Knockdown – Long-term silencing ideal for chronic disease models.
- Direct Phenotypic Insight – Establish cleargene-to-function relationships for target validation.
The core purpose of establishing shRNA libraries is to address the complexity of biological problems. It converts RNAi technology into a powerful tool for large-scale discovery, enabling scientists to systematically screen the entire genome to identify key genes linked to specific biological phenomena or disease processes, thus accelerating functional genomics research and the discovery of new drug targets.
Synbio Technologies | Custom Genome-Wide shRNA Libraries
At Synbio Tech, we provide high-quality, customizable shRNA libraries to accelerate discovery in functional genomics and drug development. Our libraries are available in multiple formats, including E. coli strains, DNA, or viral particles, tailored to your experimental needs.
- High Diversity: Our genome-wide shRNA library offers complexity from hundreds to 10⁶ unique sequences.
- High Coverage & Uniformity: Over 90% genome coverage with even sequence distribution.
- High-Quality Viral Packaging: Multiple viral systems with fast turnaround and cost-effectiveness.
- Strict Quality Control: Delivered after successful NGS sequencing validation.
Case Study
| Sample Name | XXXXX |
|---|---|
| Total Sequencing Reads | 26,115,991 |
| sgRNA Sequencing Reads | 18,659,459 |
| Reads Covered | 71.45% |
| Number of shRNAs | 11,434 |
| Covered Count | 11,110 |
| Coverage | 97.17% |
| Uncovered Count | 324 |
Reference
[1] Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014 Sep;15(9):591-600.
[2] Papadopoulos D, Ade CP, Eilers M. Generation of a pooled shRNA library for functional genomics screens. STAR Protoc. 2022 Feb 15;3(1):101183.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products