Codon optimization is one of the most important strategies to improve the expression of heterologous proteins because the use of codons varies greatly among different host cells. This can be summarized as follows: codon preference, codon coordination, and codon sensitivity. Only by understanding the secret of codon usage can we better utilize the appropriate codon to express the target protein efficiently in an experiment.
Codon Preference
Codons are redundant, but host cells do not use every codon equally to encode amino acids. A large number of studies have shown that highly expressed genes usually have a very high frequency of preferred codon use, while low expressed genes tend to random use of codons or contain a large number of rare codons. Also, the mRNA containing the preferred codon was translated faster than the artificially modified mRNA containing the rare codon. There are two hypotheses that can explain codon preference: I: mutation selection equilibrium hypothesis, which states that, under selection pressure, organisms tend to select the optimal codon to encode amino acids; II: The abundance of tRNA, the frequency of use of different codons is positively correlated with the available amount of corresponding tRNA, and the codon bias can reduce the abundance of tRNA, thus reducing the metabolic burden and is more conducive to cell growth [1].
Secondly, the use of codons can also affect the secondary structure and stability of mRNA, and affect the efficiency of protein translation. As predicted by the secondary structure of the protein, the unstructured part of the α -helix is more slowly translated. Meanwhile, hairpin structures can significantly affect protein expression, especially at the translation initiation site. Studies on 340 genomes from bacteria, archaea, fungi, plants, insects, birds, and mammals showed that, except birds and mammals, other species can improve the efficiency of translation initiation by reducing the stability of mRNA 5′- end [2].
In addition, the codon preference is also reflected in the GC content of the sequence. Studies have shown that GC content can significantly affect gene expression and regulation [3]. GC content greater than 70% will increase the stability of the RNA’s secondary structure and slow down or suspend translation. However, GC content less than 30% will slow down transcription elongation and is not conducive to protein expression. In eukaryotes, there are also some elements that have important influence on transcription, such as CpG islands, TATA boxes, and other repeat sequences which also have important influence on GC content of sequences.
Codon Coordination
The codon preference strategy can be applied to the expression of most heterologous proteins. When the amino acid sequence is preserved unchanged, the host cell preference codon is used for replacement, and the abundant tRNA in the host is used for translation, which can accelerate the translation efficiency and promote the protein expression. However, in the actual process of protein synthesis, not all codons are high-frequency codons. If the introduced codon frequency is inappropriate, the natural structure and function of the protein may be changed. Therefore, the selection of codons with appropriate frequency is a key factor for the successful expression of active protein [4]. At present, it has been reported that codon coordination optimization strategies have been successfully applied to the development and preparation of enzymes and vaccines.
Codon Sensitivity
High codon sensitivity refers to the fact that, in the absence of amino acids in cells, the binding ability of tRNA to amino acids decreases significantly with the decrease of amino acid concentration. This results in suspension or even cessation of protein expression [5]. Although the optimization strategy of codon sensitivity has not been promoted, it has important guiding significance for protein expression under extreme conditions or strict requirements on the culture medium.
Synbio Technologies NG Codon Optimization
Based on Codon preference, GC content, tandem short repeat sequences, hairpin structures, and other factors affecting the expression of heterologous proteins, Synbio Technologies independently developed our NG Codon optimization software. Our proprietary NG Codon optimization software can significantly improve the success rate of prokaryotic proteins expression and protein solubility.
Case Study 1
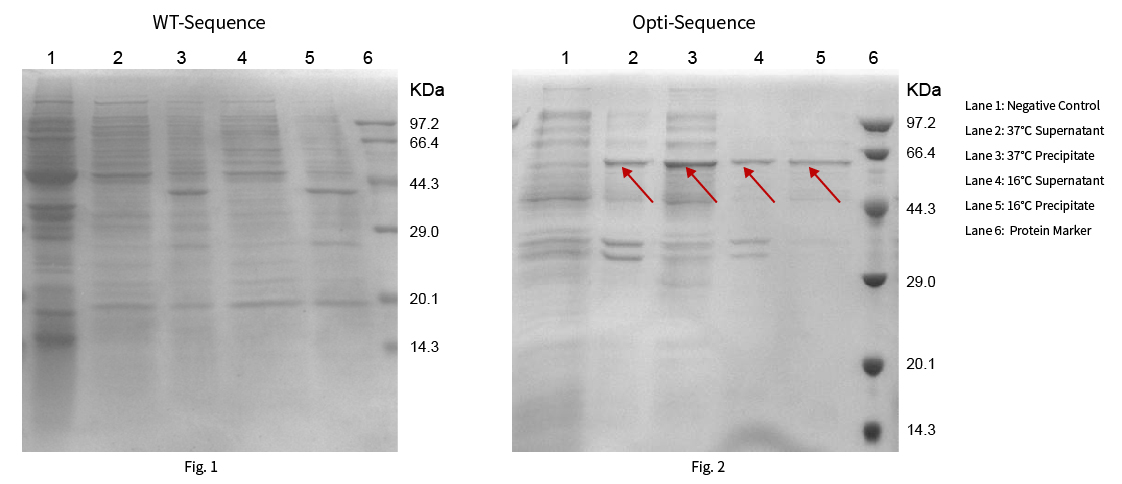
Protein R0103 has a theoretical molecular weight of 53.48kDa. The WT and optimized sequences were constructed on pET-28a(+) vector respectively, then heterologous proteins were expressed at 37 ℃ and 16 ℃. The results are shown in Fig. 1 and Fig. 2:
Results: The target protein was not expressed with the WT sequence at 37°C and 16°C. While the sequence optimized by NG™ Codon optimization software of Synbio Technologies showed significant protein expression at 37°C and 16°C.
Case Study 2
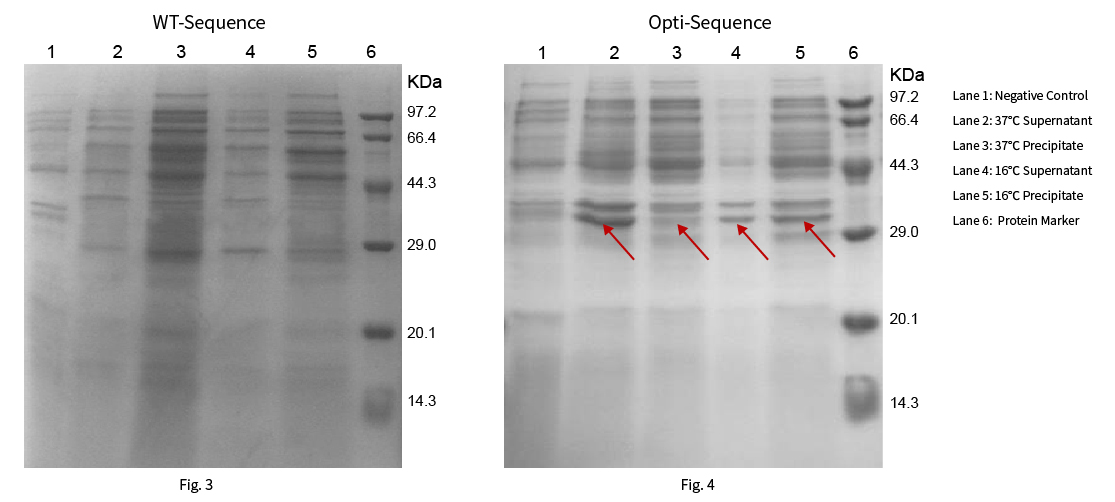
Protein R0118 has a theoretical molecular weight of 35.77kDa. The WT and optimized sequences were constructed on pET-28a (+) vector respectively, then heterologous protein expression were expressed at 37°C and 16°C. The results were shown in Fig. 3 and Fig. 4:
Results: The target protein was not expressed with the WT sequence at 37°C and 16°C. While the sequence optimized by NG Codon optimization software of Synbio Technologies showed significant protein expression and high ratio soluble protein expression at 37°C and 16°C.
References
[1] Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression [J], Trends Biotechnol, 2004, 22(7):346-353.[2] Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in Prokaryotes and Eukaryotes [J], PLoS Comput Biol, 2010, 6(2):e1000664.[3] Newman ZR, Young JM, Ingolia NT, et al. Differences in codon bias and GC content contribute
to the balanced expression of TLR7 and TLR9. Proc Natl Acad Sci USA, 2016, 113(10): E1362–E1371.[4] Angov E, Legler PM, Mease RM. Adjustment of codon usage frequencies by codon harmonization improves protein expression and folding//Evans TC Jr, Xu MQ, Eds. Heterologous Gene Expression in E. coli: Methods and Protocols. Totowa, NJ: Humana Press, 2011: 1–13.[5] Dittmar KA, Sørensen MA, Elf J, et al. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep, 2005, 6(2): 151–157.
Related Services
Protein Expression
Bacterial Protein Expression
Codon Optimization
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products