The CRISPR (Clustered regularly interspaced short palindromic repeats) system was first identified in archaea as an adaptive defensive mechanism that confers resistance to foreign genetic elements. Later on, the CRISPR-Cas system was engineered into a versatile gene-editing tool enabling manipulation of protospacer adjacent motif (PAM) downstream DNA. Now, CRISPR-Cas9 has facilitated robust genome editing in virtually any organism including: human cells, rat, mice, zebra fish, bacteria, fruit flies, yeast, nematode and etc.
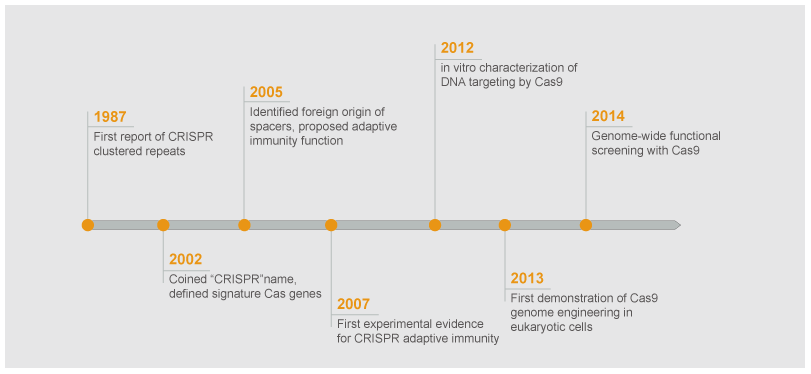
In 1987, Japanese scientists discovered a set of 29nt repeats interspaced by five intervening 32nt non-repetitive sequences in the Escherichia coli genome. The body of interspaced repeat sequences from different bacterial and archaeal strains is quickly expanding, and the nomenclature of microbial genomic loci consisting of an interspaced repeat array was unified as CRISPR (Clustered regulatory interspaced short palindromic repeats) in 2002. Over the next few years, CRISPR-Cas9 technology has been rapidly and widely adopted by the scientific community to target, edit, and modify the genomes of a vast array of cells and organisms while elucidating and refining the mechanism of CRISPR-Cas9 library genome editing. CRISPR-Cas9 has been harnessed for applications in screening for drug targets, human gene therapy, and pathogen gene disruption.
The CRISPR-Cas9 System
CRISPR is a ubiquitous family of clustered repetitive DNA elements present in 90% of Archaea and 40% of sequenced Bacteria. The 300-500bp leader located upstream of CRISPR loci is a conserved, AT-rich sequence, and is considered a promoter of CRISPR array. CRISPR array consists of repetitive sequences (repeats) interspersed with several variable sequences (spacers). Repeats are typically 21–48 nucleotides in length with the potential to form hairpin structures. The variable spacer sequences in CRISPR array are derived from previous invading mobile genetic elements (MGEs), e.g. bacteriophages and plasmids. Prokaryotes with CRISPR-Cas immune systems capture short invader sequences within the CRISPR loci in their genomes, and small RNAs produced from the CRISPR loci (crRNAs) guide Cas proteins to recognize and degrade (or otherwise silence) the invading nucleic acids. Complete genome sequencing studies showed the presence of common sequences flanking the multiple CRISPR loci in multiple prokaryotic species. Comparison of the genes that flank the CRISPR loci in the genomes of different species showed a clear homology among those genes, which was later designated as CRISPR-associated genes, Cas. Cas genes encode proteins with a variety of nucleic acid-manipulating activities such as nucleases, helicases and polymerases, and are often located adjacent to the CRISPR region.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























