Ending the Food Crisis: Unlocking the Infinite Potential of Plant Gene Editing
As the global population surges and the demand for food continues to increase, how can we minimize crop yield losses and maximize crop resilience? The advent of plant gene editing technology represents a pivotal moment, ushering in a new era for agriculture and bioscience.
CRISPR/Cas gene editing is a revolutionary technique inspired by nature. Originally derived from bacteria and archaea as a defense mechanism, this cutting-edge system allows for precise modifications to an organism’s DNA. By effectively preserving DNA fragments from viruses and external sources, CRISPR/Cas harnesses the power of both sgRNA and Cas proteins. With its intuitive operation, exceptional editing efficiency, and ability to handle multiple targets, this method has been embraced by researchers on a world-scale. Current gene editing technology brings the extraordinary potential to meticulously reshape and refine the genome of any organism.
Remarkable Achievements in Plant Gene Editing
Discover the incredible potential of CRISPR/Cas gene editing technology in enhancing crop traits.
-
Increasing Yields
By manipulating genes involved in photosynthesis, we can enhance the rate at which plants convert light energy into chemical energy, as well as improve their ability to absorb water and nutrients through their roots. This comprehensive approach results in significantly higher crop yields and greater resilience to drought conditions. -
Enhancing Disease Resistance
Shearing and inactivation of host genes, on which pathogens depend, can increase crop resistance to pathogens. By inhibiting the invasion and proliferation of pathogens, crops are far better equipped to effectively combat the threat of disease. -
Enabling Insect Resistance
By incorporating specific insect-resistant genes, crops can naturally produce proteins to repel insects. This groundbreaking method reduces the need for pesticides while safeguarding the crop from insect-related damage. -
Elevating Taste and Quality
Through the manipulation of genes associated with food quality, such as those responsible for starch synthesis and fruit ripening, crops can be modified to enhance their taste, aroma, color, nutritional value, and storage attributes – elevating their overall quality.
Tools for Plant Gene Editing
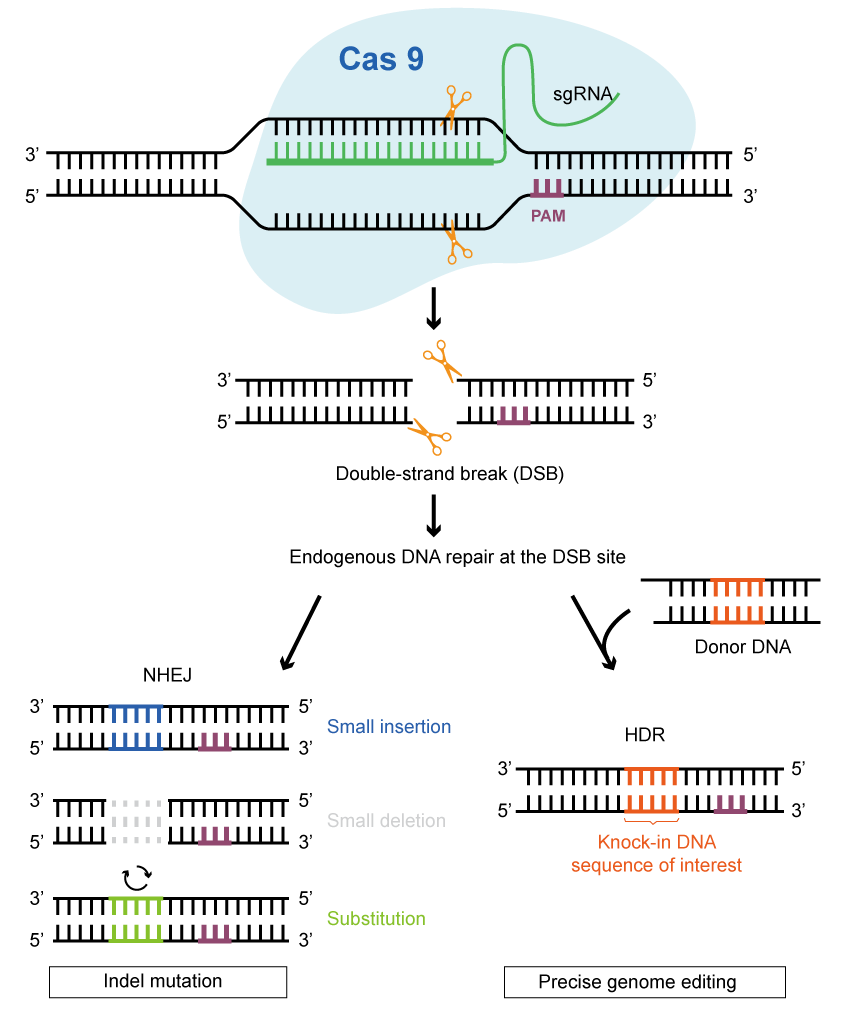
Once the Cas protein cuts the DNA to create double-strand breaks, the cell activates the DNA damage repair process to mend these breaks. The two most prevalent forms of editing are functional gene knock-out and gene fragment targeted knock-in / substitution. Moreover, single-base editing and prime editing have shown exceptional benefits in attaining higher accuracy and wider applicability.
Functional Gene Knock-out
Harnessing the power of CRISPR/Cas9 technology for the specific knock-out of functional genes, scientists have discovered its game-changing application in plant biology. This preference arises from the Cas9 protein’s ability to induce a double-strand break in the target DNA, subsequently prompting the activation of the non-homologous end-joining (NHEJ) repair pathway in the host organism. For the most part, it is possible to generate bases and insert deletions (indels) near the cut site. However, imprecise repair can result in loss of function, frameshift mutations, creation of a premature stop codon, etc.
Gene (Fragment) Targeted Knock-in or Substitution
When CRISPR/Cas9 is used to introduce a DNA double-strand break and a donor fragment at the same time, while both ends of the fragment carry sequences like those at the DNA break, then the editing acceptor has a certain probability of initiating homology-directed repair (HDR). This repair process allows for the precise insertion or substitution of the donor fragment through homologous recombination. This editing method is more accurate and versatile compared to the random insertion or deletion caused by the NHEJ pathway. By promoting the stable combination of multiple genes responsible for desirable traits, HDR overcomes the limitation of inheriting excellent traits in traditional breeding methods. As a result, it offers promising potential for a wide range of applications.
Base Editing
Single base editing technology involves the precise alteration of a single base at a specific location within a target gene fragment. The establishment of cytosine editors (CBE) and adenine editors (ABE) has enabled single-base editing to achieve four distinct types of base conversions (C→T, G→A, A→G, T→C). What sets single base editing apart is that it does not rely on creating double-strand breaks in DNA, thereby avoiding the inherent unpredictability of the NHEJ repair pathway and the constraints of the often-inefficient HDR repair pathway.
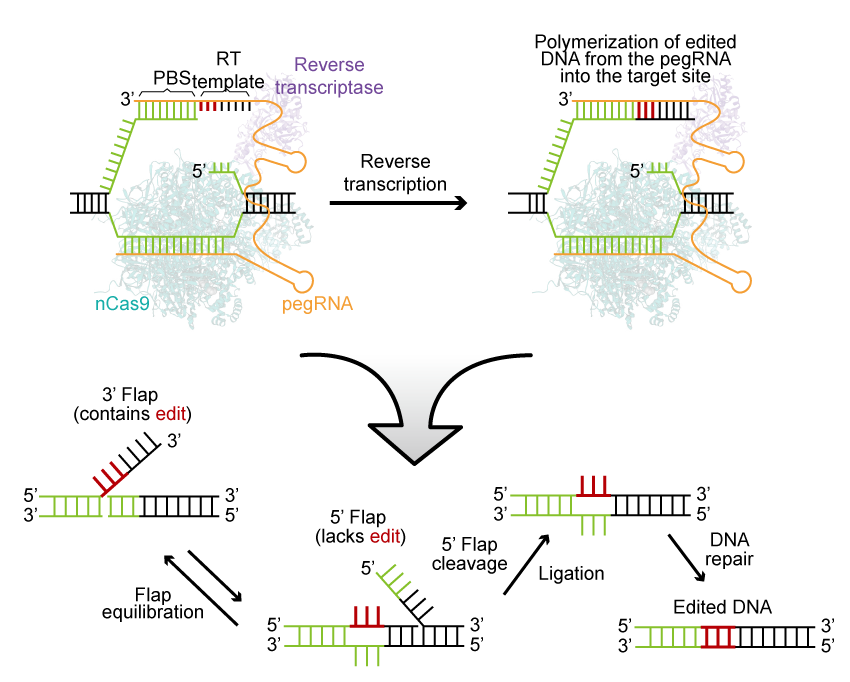
Prime Editing
Prime Editor, an ultra-precise novel DNA editing tool, facilitates a wide range of DNA editing, including substitutions, small insertions, and deletions, directly at the target site within a living cell’s genome without the need for the formation of a DSB.
Prime Editing (PE) proteins are created through the fusion of nickase Cas9 (nCas9) and reverse transcriptase (RT). These PE proteins work in conjunction with a prime-editing guide RNA (pegRNA), which serves a dual purpose. The pegRNA guides the editing protein to the target site while also providing an editable RNA template.
The process begins with pegRNA directing the PE to make a precise single-strand cleavage in the target DNA. Subsequently, it extends a new DNA sequence from the cut, using the editing template contained within the pegRNA. The cell’s endogenous repair mechanisms then automatically integrate this newly synthesized sequence into the genome.
Prime Editor stands out from other methods due to its remarkable versatility, high editing precision, and DNA target specificity.
Next-Generation Gene Editing: Paving the Way for Agricultural Innovation and Food Security with Synbio Technologies
Breakthroughs in CRISPR gene editing technology are revolutionizing the world of agriculture, presenting unparalleled opportunities to meet ever-increasing global food requirements. By allowing us to meticulously tailor the genetic makeup of crops, this groundbreaking technique holds immense potential to enhance crop productivity, fortify resistance against weather and insects, and elevate overall quality.
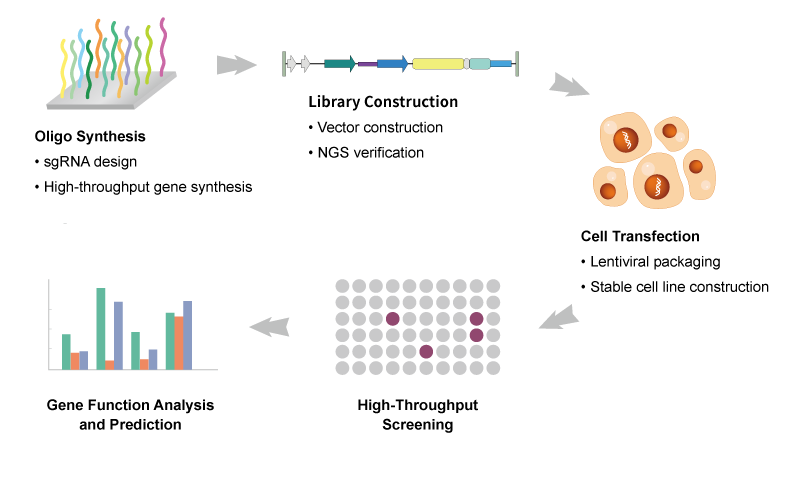
Synbio Technologies has nearly a decade of experience in whole genome editing of crops. Relying on our own intellectual property algorithms, we can provide sgRNA design and library synthesis for whole plant genomes, spanning over ten diverse species, including rice, wheat, corn, and more. Our solutions enable customers to access key genetic information pertaining to thousands of genes related to plant morphology, accelerating researchers’ exploration of genetic diversity within crops while significantly reducing the time required for crop breeding through the application of gene editing technology.
As we look to the future, plant gene editing technology is poised to continue pushing the boundaries of what is possible in the life sciences. Precise editing techniques are expected to improve, enhancing both accuracy and efficiency for researchers. Overall, this will allow for more refined control over gene regulation. Additionally, the exploration of multigene regulation will become a prominent avenue for research, offering the possibility of highly intricate improvements in plant traits.
References
[1] Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019 Dec;576(7785):149-157.
[2] Li H, Xie K. Recent progresses in CRISPR genome editing in plants. Sheng Wu Gong Cheng Xue Bao. 2017 Oct 25;33(10):1700-1711.
[3] Zhan X, Lu Y, Zhu JK, Botella JR. Genome editing for plant research and crop improvement. J Integr Plant Biol. 2021 Jan;63(1):3-33.
[4] Gao C. Genome engineering for crop improvement and future agriculture. Cell. 2021 Mar 18;184(6):1621-1635.
[5] Liu YG, Li GS, Zhang YL, Chen LT. Current advances on CRISPR/Cas genome editing technologies in plants. 2019 Vol.40 No.5 pp.38-49.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products