CRISPR-Cas9 is a revolutionary technology that can be used to edit specific genes in the human body from organisms like bacteria and viruses. The system consists of guiding proteins called sgRNA which help break apart double stranded DNA at targeted locations with Cas9 enzyme for editing purposes, leading eventually into nonhomologous end joining (NHEJ) or homologue Recombination(HR).
CRISPR-Cas9 is a game changer for many applications. It can be used in basic research to diagnose and understand diseases at the genetic level, as well as help identify drugs that will work best on those disorders. The role of sgRNA in gene editing is to accurately identify the target sequence. However, off-target effects can occur and even impact final results. Therefore, sgRNA design must be accurate for effective use during gene editing research.
sgRNA Design Factors
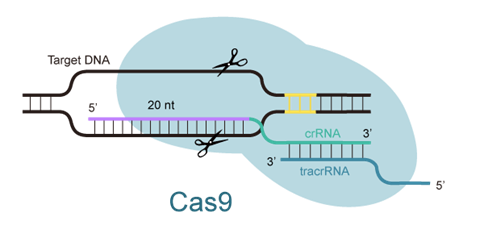
- The average length of sgRNA is ~20nt.
- The base composition of the sgRNA sequence should contain GG at the 3′ end, while the sgRNA seed sequence should avoid ending with more than 4 T’s. GC content within the sgRNA sequence should be 40%~60%.
- The number of matches between the seed sequence of the sgRNA and the off-target site must be as low as possible.
- If constructing an expression vector for U6 or T7 promoter-driven sgRNA, the 5′ base of the sgRNA needs to be considered as G or GG to improve its transcription efficiency.
- The binding position for sgRNA targeting genes is crucial for gene shift mutation. It needs as close of proximity possible with the ATG downstream from where coding occurs, preferably located in either 1st or 2nd exon.
- Check for SNPs in the genomic sequence of the sgRNA target binding site.
- If Cas9 nickase is used, the spacing of paired sgRNAs needs to be considered
- When analyzing the off-target effect of a whole gene, it is important to take into account any base mismatches at sites outside your target sequence. A minimum number of 5 bases is recommended.
The more mutations a CRISPR system has, the better. sgRNA target activity directly relates to how effectively it can cut and edit DNA in cells – leading researchers towards higher mutation efficiency for positive clones later when screening them out via RT-PCR or gene expression analysis. We have found that it is critical to design and find an active target before proceeding with gene editing. Usually, after designing a specific targeting sequence for your desired product(s), in vitro cellular activity screening needs be done to ensure effective targets can successfully receive modifications or deletions from the Cas9 enzyme .
sgRNA Design | Synbio Technologies
CRISPR-Cas9 is a new and innovative technology that is considered to be the most promising tool in gene modification, based on its precise editing of the genome. This process utilizes double stranded DNA (dsDNA) breaks at specific loci, which can be induced by sgRNA to recognize target sites on chromosomes.
Synbio Technologies offers professional sgRNA design services to help researchers accelerate their CRISPR-based gene editing projects. Our team of experienced scientists has a proven track record in designing highly efficient sgRNAs that achieve the desired editing outcomes. We utilize a variety of state-of-the-art platforms and algorithms to select the most effective sgRNA sequences for your target gene. In addition, we also offer customized sgRNA libraries to suit your specific needs. Our services are fast, reliable, and affordable, and we guarantee 100% satisfaction.
Contact us today to learn more about our sgRNA design services!
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























