The 2024 Nobel Prize in Chemistry is awarded to David Baker, Demis Hassabis, and John M. Jumper for their outstanding contributions to the fields of protein design and structure prediction.
David Baker: Pioneering Achievements in Computational Protein Design

David Baker. Ill. Niklas Elmehed © Nobel Prize Outreach
David Baker's contribution lies in his successful development of the Rosetta software, which designs new proteins that do not exist in nature. His innovative achievements include:
· Rosetta Enzymes: Enzymes that can catalyze specific chemical reactions.
· Mini-proteins: Small proteins designed for specific biological functions.
· Protein-based Nanomaterials: Proteins that can be used to construct novel nanomaterials.
Baker's pioneering achievements in computational protein design have not only pushed the boundaries of protein science but also brought revolutionary breakthroughs in drug development, materials science, and environmental management.
David Baker was born in 1962 in Seattle, Washington. He is currently the Director of the Protein Design Institute at the University of Washington and a prominent researcher at the Howard Hughes Medical Institute. His research has garnered significant attention, and he was awarded the Science Breakthrough Prize in Life Sciences in 2021.
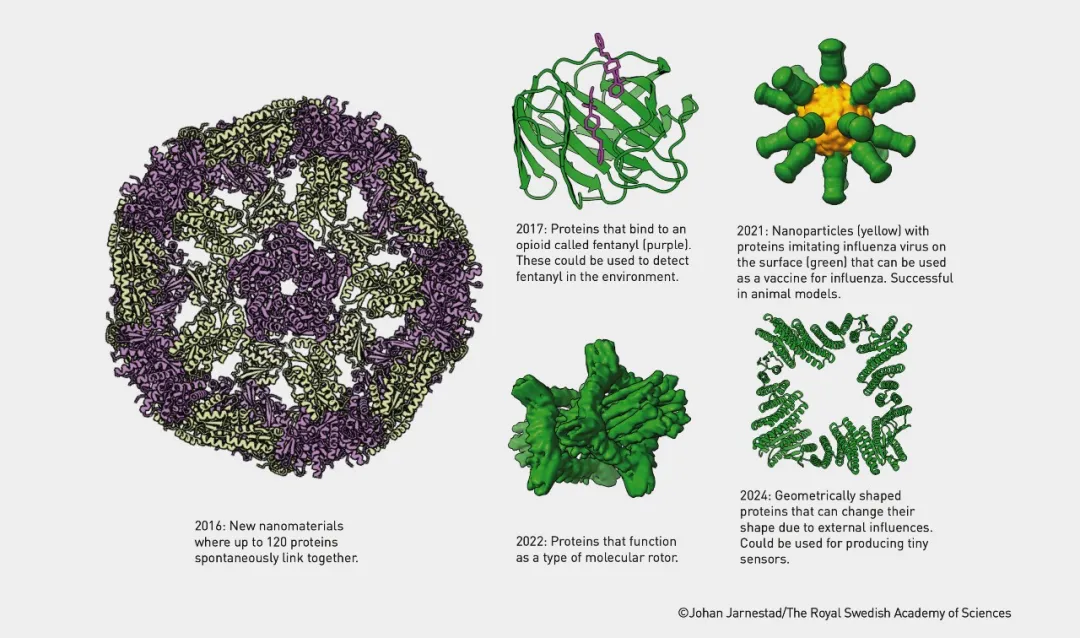
Constructing proteins using Rosetta software © Johan Jarnestad/The Royal Swedish Academy of Science
Demis Hassabis and John Jumper: AI Models for Predicting Protein Structure
-

Demis Hassabis. Ill. Niklas Elmehed © Nobel Prize Outreach
-

John Jumper. Ill. Niklas Elmehed © Nobel Prize Outreach
Demis Hassabis and John Jumper received shared honors for their achievements in protein structure prediction. Their AI model tool, AlphaFold, successfully addressed the long-standing challenge of predicting protein structures, which has perplexed the scientific community for over 50 years. AlphaFold can predict the three-dimensional structure of proteins directly from their amino acid sequences, achieving an accuracy exceeding 90%, far surpassing its competitors.
Since its initial success in the CASP competition in 2018, the DeepMind team's AlphaFold 2, released in 2020, has revolutionized the field of protein structure prediction. In 2021, they publicly released predicted structures for nearly 98.5% of human proteins. In 2024, AlphaFold 3 further enhanced prediction accuracy, covering nearly all biomolecules, including proteins, DNA, RNA, and their interaction patterns.
Demis Hassabis, founder of DeepMind, is an outstanding scientist with a background in computer science and neuroscience. Born in London in 1976, he graduated with a degree in Computer Science from Cambridge University and later earned a Ph.D. in Cognitive Neuroscience from University College London, followed by postdoctoral work at MIT and Harvard.
John Jumper, a senior research scientist at DeepMind and the first author of AlphaFold, specializes in using machine learning to simulate protein folding and dynamics. Born in 1985 in Arkansas, he majored in theoretical physics during his undergraduate studies and later earned a Ph.D. from the University of Chicago.
Their research not only advances the field of protein science but also opens new avenues for drug design and vaccine development. Previously, the DeepMind team made headlines with AlphaGo, which defeated world champion Lee Sedol at the 2016 World Go Championship, marking a significant breakthrough in artificial intelligence in the realm of board games.
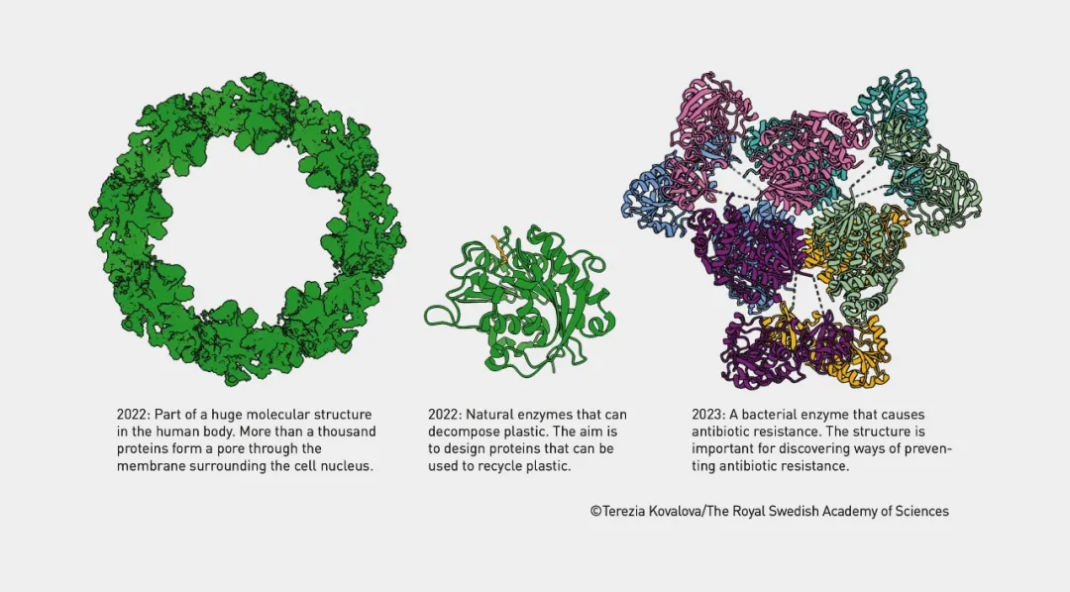
Designing protein structures with AlphaFold2 ©Terezia Kovalova/The Royal Swedish Academy of Science
Future Outlook
The research achievements of David Baker, Demis Hassabis, and John Jumper exemplify the importance of scientific innovation and interdisciplinary collaboration. Although significant progress has been made in the field of protein structure prediction, challenges remain, including structural complexity and biological diversity.
We extend our deepest respect to the Nobel Prize winners in Chemistry and express our gratitude for their contributions to protein design and structure prediction, eagerly anticipating further breakthroughs in scientific research!
References:
1. Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, Kleywegt GJ, Bateman A, Evans R, Pritzel A, Figurnov M, Ronneberger O, Bates R, Kohl SAA, Potapenko A, Ballard AJ, Romera-Paredes B, Nikolov S, Jain R, Clancy E, Reiman D, Petersen S, Senior AW, Kavukcuoglu K, Birney E, Kohli P, Jumper J, Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021 Aug;596(7873):590-596. doi: 10.1038/s41586-021-03828-1.
2. Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024 Jun;630(8016):493-500. doi: 10.1038/s41586-024-07487-w.
3. https://www.nobelprize.org/prizes/chemistry/2024/press-release/
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























