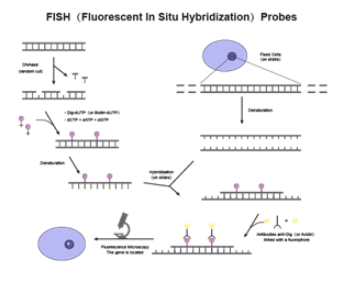
Fluorescence In Situ Hybridization (FISH) is a technique that combines the high sensitivity, safety, and visualization of fluorescent signals with the high specificity of in situ hybridization. It employs fluorescently labeled nucleic acid probes that hybridize to target nucleic acids in samples. Fluorescent signals are then identified and quantified under a fluorescence microscope, enabling the detection and diagnosis of chromosomal or genetic abnormalities in cells and tissue samples. This provides accurate information for the classification, prognosis, and treatment of various gene-related diseases.
Types of FISH Probes
Chromosome-Specific Repetitive Sequence Probes: These probes bind to specific chromosomal structures and recognize repetitive DNA sequences. For example, α-satellite and satellite III probes target hybridization loci typically larger than 1 Mb, without scattered repetitive sequences. They bind tightly to the target site, producing strong hybridization signals that are easy to detect.
Whole Chromosome or Chromosome Region-Specific Probes: Composed of distinct nucleotide fragments from an entire chromosome or a specific chromosomal region, these probes are derived from large chromosome-specific DNA fragments cloned into phages or plasmids. They are typically used in metaphase chromosome analysis to identify marker chromosomes or structural abnormalities.
Single-Sequence Probes: These probes hybridize to single-copy DNA sequences within specific chromosomal regions or genes. Common types include locus-specific probes and gene-specific probes.
Applications of FISH Probes
1. Cancer Diagnosis
-
Hematologic Oncology: FISH is widely used in the diagnosis of blood cancers, focusing on detecting fusion genes resulting from chromosomal translocations and identifying key gene deletions critical for disease diagnosis and prognosis. FISH can also monitor minimal residual disease and the status of hematopoietic stem cell transplantation.
-
Solid Tumor Oncology: While Next-Generation Sequencing (NGS) and PCR are common for detecting gene mutations, they have limitations. NGS requires sophisticated lab setups, has longer turnaround times, and is costly. Fluorescent PCR is limited to relative quantification using standard curves, while digital PCR has high system costs, limited throughput, and complex operations. Additionally, they lack the ability to localize gene mutations at the cellular level. In contrast, FISH provides direct evidence of DNA amplification within interphase nuclei, with fluorescence signal intensity and quantity correlating with amplification levels.
/li>
2. Gene Mapping
FISH's high sensitivity and ability to detect multiple genes simultaneously make it valuable for determining the precise location of target genes, the relationships between multiple genes, and the positioning of genes relative to chromosomal structures such as telomeres and centromeres. It is essential for constructing gene maps and is widely used in physical gene mapping and genomic studies.
FISH probes play a crucial role in prenatal diagnosis by detecting chromosomal abnormalities in fetal cells, such as numerical or structural anomalies. This enables the timely identification and diagnosis of chromosomal disorders like Down syndrome, trisomy 18, and trisomy 13, providing valuable screening and diagnostic services to minimize the impact of chromosomal disorders on newborns.
4. Genetic Disease Diagnosis
FISH probes can detect gene mutations or deletions associated with genetic diseases. For instance, specific FISH probes can determine whether certain genes on chromosomes are abnormal, aiding in diagnosing genetic disorders. This is particularly valuable for diseases caused by chromosomal abnormalities or single-gene defects, such as Down syndrome or specific leukemias.
5. Viral Infection Detection
FISH probes can also be used to detect viral infections by identifying the localization of viral nucleic acids within host cells, helping to determine the status and severity of the infection.
What is the difference between qPCR probe and FISH probe?
|
|
qPCR |
FISH |
|---|---|---|
|
Specificities |
Real-time monitoring with high accuracy and sensitivity |
High specificity, high sensitivity, visualization |
|
Target of Detection
|
The amplification product of DNA or RNA |
DNA or RNA sequences in cell or tissue sections |
|
Resolution
|
Quantification of the concentration of DNA or RNA by changes in the intensity of the fluorescent signal with high resolution. |
Capable of detecting genomic variants at the individual cell level with very high resolution. |
|
Application |
Gene expression analysis, pathogen detection, gene mutation detection, DNA methylation analysis, etc. |
Chromosome variant testing, genetic disease diagnosis, tumor diagnosis and prognosis evaluation, prenatal diagnosis, etc.
|
Synbio Technologies |Diagnosis Probe Services
Synbio Technologies offers standard oligos, fluorescent probes, special modifiers, skeleton modifiers, oligo pools, and other service lines to provide customers with a variety of customized FISH probe raw materials.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























