In recent years, the field of synthetic biology has seen incredible advancements in the realm of digital data storage. Researchers have been working tirelessly on finding ways to store vast amounts of information using the basic building blocks of biology. These efforts have been met with great success, leading to the creation of new methods and techniques that have the potential to revolutionize the way we store and access data.
At the forefront of this exciting development is the concept of DNA-based data storage. This technology involves encoding digital information into the four basic building blocks of DNA, known as adenine, thymine, guanine, and cytosine. These building blocks, or nucleotides, are arranged in a specific sequence to create what is known as a DNA strand. Once encoded, this strand can be synthesized and stored in a physically small space, making it an incredibly efficient method of data storage.
But the advantages of DNA-based data storage go beyond just its compact size. Due to the robustness and stability of DNA molecules, the information encoded using DNA has the potential to last for thousands of years without the need for maintenance or repair. Additionally, DNA data storage is incredibly dense, with a single gram of DNA capable of storing as much data as a million CDs.
However, the challenge of DNA-based data storage lies in the ability to read and retrieve the encoded information. Traditional methods of data retrieval, such as reading from hard drives or solid-state drives, are not applicable when it comes to DNA. Instead, researchers have developed techniques such as next-generation sequencing and polymerase chain reaction (PCR) to read DNA-encoded data.
Excitingly, the potential of DNA-based data storage goes beyond just data centers and personal computers. In recent years, researchers have also been exploring the possibility of using DNA as a way to store information in living organisms. By encoding information into the DNA of living organisms, it may be possible to create biological systems that can store and transmit data in ways we previously thought impossible.
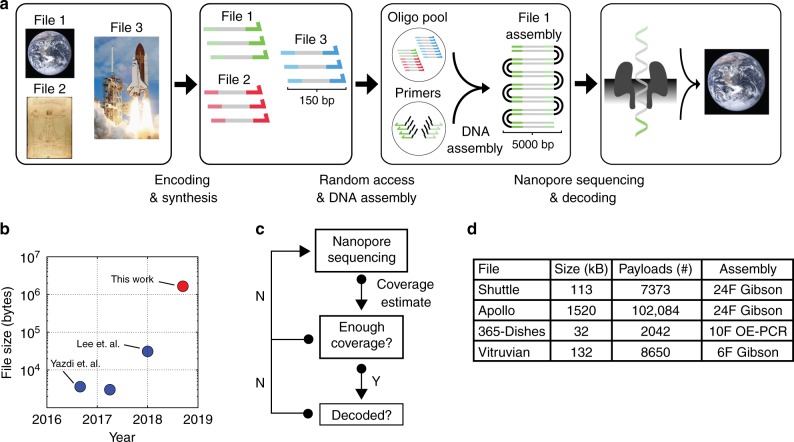
This paper reports a method that combines random access, DNA assembly, and nanopore sequencing to decode the information stored in DNA. Multiple digital files were mapped into 150 nucleotide DNA sequences and synthesized. Gibson assembly and Overlap-Extension PCR (OE-PCR) successfully decoded 1.67MB of digital information stored in DNA. Nanopore sequencing can estimate the decoding coverage of digital files stored in DNA in real time. Four different DNA coding files were amplified, assembled, and sequenced using the ONT MinION platform. An assembly of 6, 10, or 24 fragments were built for each file. By storing 1.67 megabytes of digital information in 111,499 oligonucleotides, the ability of sequencing and decoding has been improved by two orders of magnitude, and a faster and more flexible DNA storage scheme has been developed.
Fig.1 Overview of the DNA data storage workflow (Lopez, 2019)
Overall, the field of DNA-based data storage represents a fascinating frontier in the world of synthetic biology. With the potential to store massive amounts of data in a compact and stable form, and the possibility of using living organisms as data storage devices, the future of data storage is looking more exciting than ever before.
Gene Synthesis and DNA Data Storage | Synbio Technologies
Synbio Technology has a complete synthesis platform, providing services such as DNA fragment synthesis, gene synthesis, pathway synthesis and assembly, and genome synthesis and assembly. We have rich experience in complex and long gene synthesis. We guarantee the delivery of 100% accurate gene sequences, so that you can obtain high-quality products with minimal project funds.
We have also developed DNA Studio to achieve bi-directional transcoding between “A, T, C, G ” and digital information. Then, according to the code, DNA sequences are synthesized accurately on chips, rapidly as well as massively. We invite you to join in developing DNA digital storage systems.
Reference
Lopez, R., et.al (2019). DNA assembly for nanopore data storage readout. nature communications. https://doi.org/10.1038/s41467-019-10978-4
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products