Did you know that up to 50% of protein expression experiments fail due to common yet avoidable mistakes? Protein expression is a cornerstone of life sciences research and biopharmaceutical development, but unsuccessful attempts can be frustrating and costly. Here, we explore five common protein expression challenges and provide actionable solutions to overcome them.
1. Codon Mismatch
Codon mismatch is a common yet often overlooked cause of protein expression failure. Different organisms prefer specific codons during protein translation. If your gene contains rare codons that the host organism doesn't frequently use, translation can stall, leading to poor protein yields. This issue is especially problematic in high-expression systems, where a shortage of tRNA for rare codons can halt translation altogether.
Solution:
Codon optimization is the key to overcoming this challenge. By modifying the gene sequence to include codons that are more common in your host organism, you can significantly improve translation efficiency. Modern gene synthesis technologies make it easy to optimize the gene sequence based on the tRNA usage frequency of the host. For example, in E. coli, selecting more frequent codons can speed up translation and boost protein yields.
2. Incorrect Protein Folding and Inclusion Body Formation
In high-expression systems, especially in prokaryotic systems like E. coli, proteins often fail to fold correctly, forming inclusion bodies—insoluble aggregates of misfolded proteins. This is a particular concern for complex proteins or those requiring disulfide bonds.
Solution:
-
Lowering expression temperature: Slowing down translation by reducing the temperature (e.g., 20–30°C) gives proteins more time to fold correctly.
-
Fusion tags: Using tags like GST or MBP can enhance protein solubility and prevent misfolding.
-
Co-expressing chaperones: Co-expressing molecular chaperones, such as GroEL/GroES or DnaK/DnaJ, can assist in correct protein folding and prevent inclusion body formation.
3. Incorrect Expression System
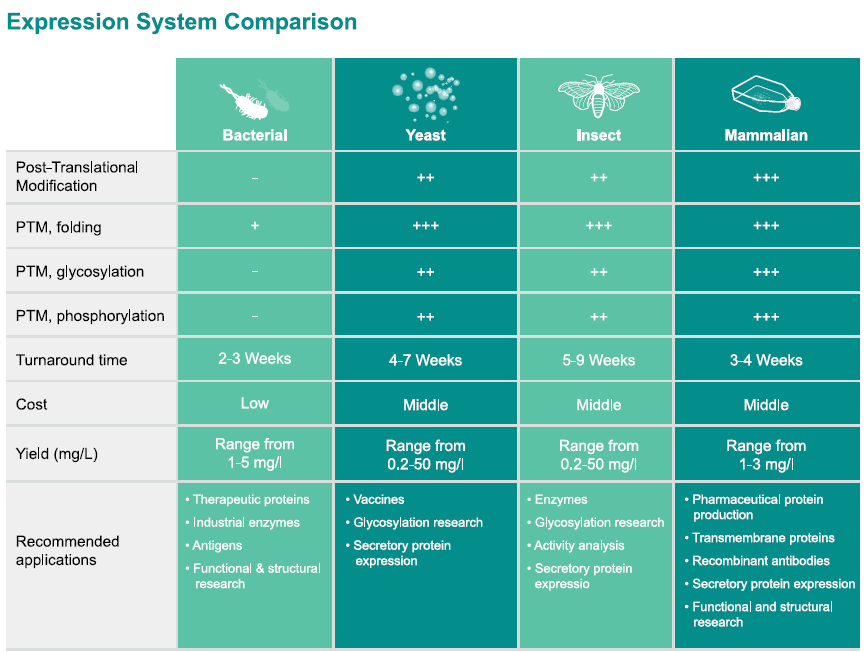
Not all proteins are suited for all expression systems. Some proteins, especially eukaryotic ones, require post-translational modifications (e.g., glycosylation or phosphorylation) that prokaryotic systems like E. coli cannot provide, leading to inactive or nonfunctional proteins.
Solution:
Choose the right expression system based on your protein’s needs:
-
For eukaryotic proteins:Use yeast, insect cells, or mammalian cells, as these systems can perform necessary modifications like glycosylation.
-
Optimize the vector: Choose the right promoter (e.g., T7 for high expression) and selection markers to maximize yield and functionality.
-
Consider hybrid systems: If cost is a concern, explore systems likePichia pastoris (yeast), which combines eukaryotic modification capabilities with prokaryotic ease of use.
4. Toxic Protein Expression
Expressing certain proteins, such as complex enzymes or viral proteins, can be toxic to host cells. This toxicity can stunt cell growth or even lead to cell death, drastically reducing protein yields and complicating your experiments.
Solution:
-
Use inducible expression systems: Use tightly controlled expression systems (e.g., lac operon or arabinose-inducible systems) to express the protein only under specific conditions, minimizing its toxic effects on the cells.
-
Switch to low-copy plasmids: These plasmids slow down protein production, preventing overload and giving cells time to cope with the toxic protein.
-
Optimize expression timing: Induce protein expression during the log phase of cell growth, when cells are most robust and can better handle stress.
5. Protein Degradation
Protein degradation is often due to proteases within the host cell recognizing degradation signals in the protein, or due to structural instability of the protein itself. Especially for structurally unstable proteins, they are susceptible to degradation by endogenous proteases of the host.
Solution:
-
Use protease-deficient strains: Strains like BL21(DE3) lack specific proteases, reducing the risk of degradation during expression.
-
Add protease inhibitors: Including inhibitors during purification can prevent degradation during extraction and storage.
-
Protein engineering: Removal of readily recognized degradation signals or introduction of mutations to enhance protein stability, thereby reducing degradation.
Synbio Technologies | Protein Engineering Services
AtSynbio Technologies, we understand the complexities of protein expression. Our comprehensive solutions are designed to tackle the challenges listed above and more.
Highlights
1. One-Stop Protein Expression Solution:From Codon optimization and AI-drivengene synthesis to vector selection, expression system screening, protein purification, and functionality validation.
2. Multiple Expression Systems: We offer a wide range of expression systems, including E. coli, yeast, insect cells, and mammalian cells, allowing us to tailor solutions for your specific needs.
3. Customized Optimization Strategies: We work closely with you to design the most effective strategies for your protein expression project, considering the unique requirements of your target protein.
4.High Success Rate: With years of industry experience, our optimized protocols and expertise dramatically increase your chances of successful protein expression.
References
1. Gustafsson, C., Govindarajan, S., & Minshull, J. (2004). Codon bias and heterologous protein expression. *Trends in Biotechnology*, 22(7), 346-353.
2. Rosano, G. L., & Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. *Frontiers in Microbiology*, 5, 172.
3. Baneyx, F., & Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. *Nature Biotechnology*, 22(11), 1399-1408.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























