The Evolution of De Novo DNA Synthesis
The history of oligo synthesis dates back to the 1950s, with early methods involving phosphodiester, H-phosphonate, and phosphotriester approaches. Today, the dominant chemistry for oligo synthesis has evolved, and we now have access to high-throughput array-based methods that enable the synthesis of large DNA constructs at low costs. This technological advancement has been crucial in driving the field of synthetic biology forward.
Over the past few decades, advancements in De Novo DNA synthesis have revolutionized the field of synthetic biology. Unlike traditional methods that rely on the modification of existing DNA sequences, De Novo synthesis allows researchers to create entirely new genetic sequences from scratch. This technology has opened up a vast array of emerging applications, ranging from the development of novel therapeutics to the creation of synthetic genomes.
Development of Synthetic Genomes
By synthesizing entire genomes, researchers can gain complete control over an organism's genetic code. This has led to significant progress in the field of viral genomic reconstructions, particularly for RNA viruses. Synthetic versions of viruses such as poliovirus and influenza have been created for purposes such as viral attenuation, vaccine development, and historical reconstructions.
Beyond viral genomes, synthetic biology researchers have also designed and built fully synthetic bacterial genomes. The Venter Institute, for example, successfully designed, built, assembled, and transplanted a fully synthetic genome into a bacterial cell. This achievement demonstrates the potential of De Novo DNA synthesis to create entirely new organisms with tailored genetic properties.
Engineering of Genetic Networks and Metabolic Pathways
Synthetic biology is heavily focused on building and optimizing genetic networks to control cellular behavior and metabolic pathways for chemical production. While many efforts are directed at assembling existing DNA in various combinations, De Novo synthesis provides a powerful tool to generate the requisite system components.
By synthesizing new genetic sequences, researchers can engineer genetic networks and metabolic pathways with unprecedented precision. This has led to the development of novel biotechnological applications, such as the production of biofuels, pharmaceuticals, and other chemicals. For instance, De Novo synthesis has been used to create synthetic metagenomics libraries for part mining, allowing researchers to identify and utilize orthogonal repressors and transcription factors to fine-tune genetic networks.
Protein Engineering and Functional Genomics
Protein engineering has always benefited from improvements in synthetic capabilities, such as DNA shuffling, site-directed mutagenesis, and low-cost gene synthesis. However, De Novo synthesis offers a more powerful tool to engineer new protein functions. By synthesizing entirely new genetic sequences, researchers can create proteins with novel properties and functions.
This technology has significant implications for functional genomics, as it allows researchers to test hypotheses about the structure-function relationships of proteins in a high-throughput manner. By synthesizing large libraries of synthetic genes and using next-generation sequencing (NGS) to measure their functional consequences, researchers can rapidly identify and characterize new protein functions.
Creation of Synthetic Gene Libraries
Synthetic Gene Libraries offer versatile applications, including targeted capture and resequencing of exons and other genomic regions of interest. By synthesizing large pools of oligos and assembling them into longer double-stranded DNA constructs, researchers can generate comprehensive libraries of synthetic genes that cover the entire genome of an organism.
These libraries also have significant implications for personalized medicine and precision genomics. By sequencing an individual's genome and comparing it to the synthetic gene library, researchers can identify genetic variants and mutations that are associated with disease. This information can then be used to develop targeted therapies and personalized treatment plans.
Data Storage in DNA
Recently, the potential of DNA as a high-density data storage medium has been demonstrated by researchers. They digital information into DNA sequences, enabling vast data to be stored in a compact and durable format. De novo DNA synthesis allows researchers to synthesize large pools of oligos for digital information encoding.
This technology has significant implications for data storage and archiving. In contrast to traditional storage media prone to degradation and loss, DNA-based data storage is highly durable, lasting thousands of years. This makes it an ideal solution for archiving important data, such as historical records, medical records, and scientific data.
DNA Origami and Nanotechnology
De Novo DNA synthesis has enabled the development of DNA origami, a technique for folding DNA into complex nanoscale shapes and patterns. By synthesizing long single-stranded DNA oligos and using them as scaffolds, researchers can create intricate three-dimensional structures with nanometer-scale precision.
These DNA origami structures have significant implications for nanotechnology and materials science. They serve as templates for the assembly of other nanoscale materials, like carbon nanotubes and quantum dots. Additionally, they can be modified with functional groups to create novel materials with unique properties, such as conductivity, magnetism, and bioluminescence.
Challenges and Opportunities
Although large de novo DNA synthesis has numerous benefits, several challenges remain. One of the main challenges is the error rate during oligo synthesis and assembly. High error rates can lead to functional defects in the synthetic genes and pathways, making it difficult to achieve the desired outcomes. Additionally, the expense of gene synthesis can constrain certain research endeavors. With the continuous technological progress and the growing need for synthetic DNA, there is a significant likelihood of cost decreases in the future.
Another opportunity lies in the development of new tools and methods for the design and optimization of synthetic genes and pathways. With the increasing availability of computational resources and bioinformatics software, researchers can now generate powerful statistical hypotheses for how genome sequence controls cellular functions across organisms and populations. These hypotheses can then be tested experimentally using synthetic DNA constructs.
Conclusion
Large De Novo DNA synthesis has opened up a vast array of emerging applications in synthetic biology, protein engineering, functional genomics, data storage, and nanotechnology. By synthesizing entirely new genetic sequences, researchers can create organisms with tailored genetic properties, engineer novel protein functions, generate comprehensive synthetic gene libraries, store vast amounts of data in a compact and durable format, and create intricate nanoscale structures with unprecedented precision. As this technology continues to advance, we can expect to see even more exciting and innovative applications in the future.
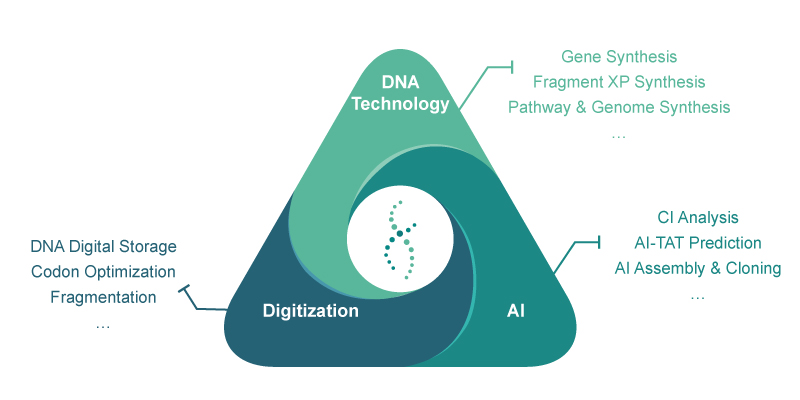
Syno GS Platform | Synbio Technologies
At Synbio Technologies, we provide a one-stop solution for DNA synthesis with a complete large De Novo DNA synthesis platform. The Syno GS platform has a variety of intelligent biological analysis tools, including Syno Ab, NG Codon, Complexity Index ( CI ) and AI-TAT. You only need to provide the nucleotide or amino acid sequence you want to synthesize, we will provide 100 % accurate gene sequence, and cloned into your designated vector. Accurate gene sequences can improve the accuracy and efficiency of protein expression.
Reference
[1] Kosuri S, Church GM. Large-scale De Novo DNA synthesis: technologies and applications. Nat Methods. 2014 May;11(5):499-507.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























