The difficulty of a DNA sequence often lies not in its length, but in the intricate arrangement of its bases. When a sequence is filled with repetitive segments, it becomes one of the toughest challenges for gene synthesis experts.
Today, we’re proud to share a groundbreaking success story: faced with an exceptionally repetitive 9 kb gene labeled by analysis software as “extremely complex,” we managed to shorten the traditional three-month synthesis timeline to just one month.
An “Extremely” Complex Gene Sequence
We received the original sequence provided by our client. Professional bioinformatics analysis revealed the Challenge behind the “Extreme” and “Complex” labels on the report: the sequence was approximately 9 kb in length—a substantial size in itself.
The true difficulty, however, lay in its high level of repetitiveness. There were 293 instances of long repeats, which can be up to 20 bases in length, classified as “Extreme.” In addition, short repeats of 9 bases accounted for 65.3% of the total sequence, meaning that nearly two-thirds of it was composed of repetitive regions.
In other words, we were dealing with a sequence that exhibited a high degree of structural self-similarity, posing exceptional challenges for accurate synthesis and assembly.
From “Sequence Optimization” To “Efficient Synthesis”
Faced with such a challenge, we completed the project in two key phases:
Phase 1: Intelligentdesign —reducingcomplexity at thesource
Our first task was to perform a “refined reconstruction” of the sequence without altering the encoded amino acid sequence. Using our proprietary sequence optimization algorithm, we restructured the original sequence to minimize synthesis difficulty.
-
Significantly reduced long repeats: The number of 20 nt long repeats was lowered from 293 to 114.
-
Optimized short repeat structures:The sequence was reorganized to effectively disperse short-repeat density and disrupt repeat continuity.

Although the optimized sequence remained functionally identical to the original, its synthetic accessibility improved dramatically—paving the way for successful laboratory synthesis.
With the optimized sequence in hand, we launched a highly efficient synthesis process. Through a combination of segmental synthesis and precision assembly, reinforced by multiple rounds of rigorous quality control, we ensured accuracy at every step.
As a result, we successfully synthesized the full-length 9 kb high-repetition gene within just one month. Sequencing verification confirmed that the product was 100% correct, with no mutations or deletions, ready for direct use in downstream expression experiments.
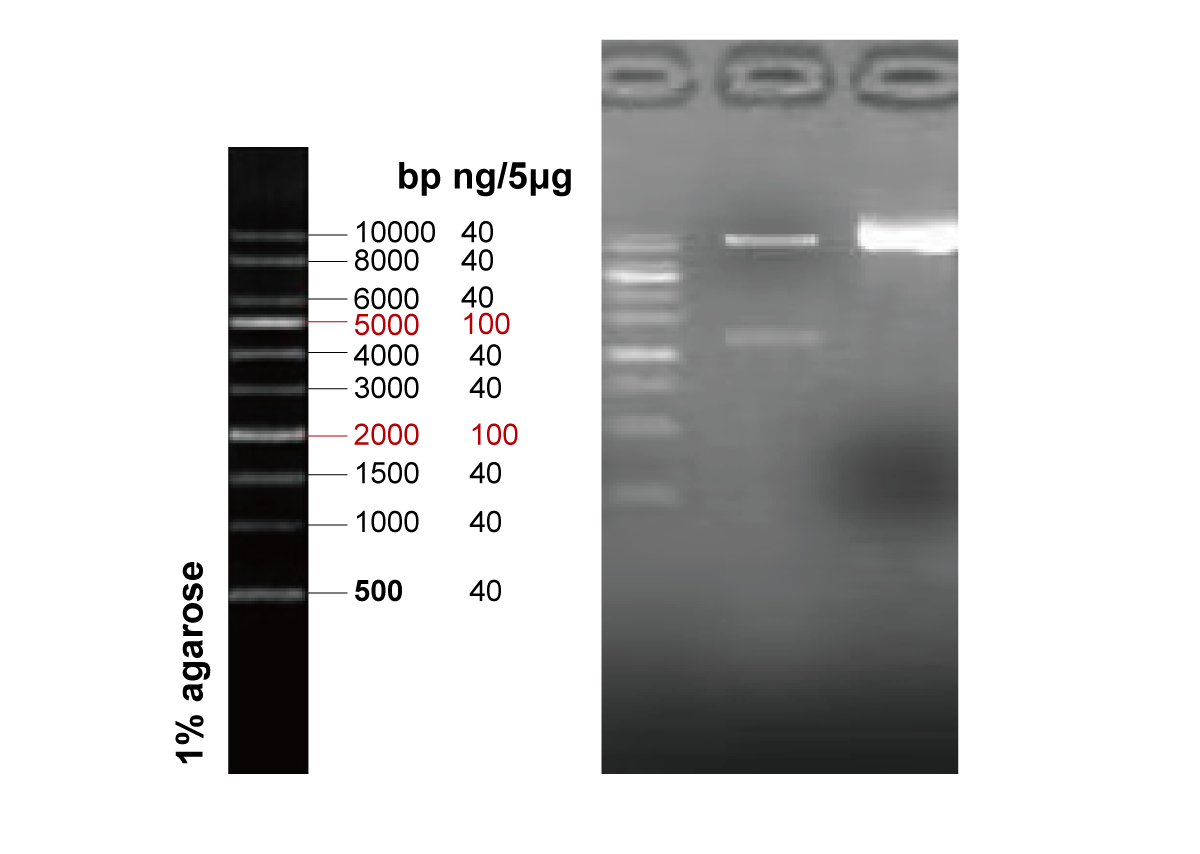
Why Repetitive Sequences Are A Nightmare For Gene Synthesis
Simply put, repetitive sequences refer to identical or highly similar base patterns that appear multiple times along a DNA strand. They’re like long sentences or phrases repeated over and over again in a text. In this case, we mainly dealt with two types:
Short repetitive sequences: Usually 6-15 bases long, these affect the specific binding of primers during DNA synthesis.
Long repetitive sequences: Generally longer than 15 bases, these are the primary disruptors during gene assembly.
Why Are Repetitive Sequences So Difficult To Synthesize?
1. Primer misalignment leading to PCR failure
During synthesis, primers act as “locators” to amplify or assemble DNA fragments. When a sequence contains numerous repeats, primers may fail to find a unique binding site and instead attach to multiple similar regions. This can generate incorrect or variable-length products—or even prevent the target fragment from amplifying at all.
2. Misassembly during construction
Gene synthesis typically involves assembling short DNA fragments step by step, much like putting together a puzzle. When too many pieces look the same, the assembly system can “get confused,” making it difficult to determine the correct order. Long repetitive sequences often cause fragment misalignment and recombination errors, resulting in structurally disordered final products.
3. Polymerase slippage
When DNA polymerase replicates repetitive regions, it is prone to “slippage.” The enzyme can lose track within the repeat, causing base insertions or deletions that introduce mutations—making the final sequence deviate from the intended design.
4. Formation of complex secondary structures
Repetitive sequences tend to fold back on themselves, forming stable secondary structures such as hairpins or stem-loops. These structures can physically obstruct the progress of synthesis enzymes, drastically reducing efficiency or even halting the reaction.
Because these factors compound one another, synthesizing highly repetitive sequences becomes an arduous process—often requiring extensive troubleshooting and long production times.
Delivering On Our Promise With Technical Excellence
Completing this 9 kb repetitive gene project ahead of schedule highlights our strengths in sequence design, synthesis efficiency, and execution.
At Synbio Technologies, we provide end-to-end solutions covering every step from codon optimization and gene synthesis to vector construction, protein expression, and purification,ensuring high expression levels and superior yields of target proteins.
We are committed to turning the “impossible” into the “achievable,” and transforming “long waits” into “rapid delivery.”
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries Genome KO Library
Genome KO Library Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard Genome KO Libraries
Standard Genome KO Libraries Standard Genome Editing Plasmid
Standard Genome Editing Plasmid ProXpress
ProXpress Protein Products
Protein Products
























