Plasmids are very small, circular DNA molecules found in organisms such as bacteria and yeast that are capable of autonomously replicating independent of the cell chromosome. In genetic engineering, plasmid preparation allows for plasmids to be used as a vector for the target gene to be expressed normally in a new host organism.
Plasmid Preparation Steps
After PCR amplification of the exogenous DNA, the plasmid vector and the exogenous DNA fragment are cut with restriction endonucleases, respectively. Next, the DNA fragment is connected to the plasmid vector via DNA ligase and then transferred to the host bacteria. Lastly, the recombinant clone is obtained through screening and identification.
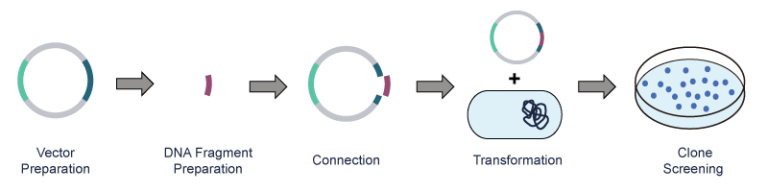
Key Technologies of Plasmid Preparation
1. PCR amplification
1) In the progression of PCR amplification, dimers and non-specific bands (size errors) frequently occur as copy number increases. Amplification specificity can be improved by reducing the concentration of the template, primers, and magnesium ions, reducing the amount of enzymes, and increasing the annealing temperature. The quality of primer design is key. In addition to following traditional primer design rules, enzymatic cleavage sites and protective bases are often added to the primer sequence when constructing the vector.
2) If the amplified fragments appear to diffuse on the gel, the ratio of each component in the reaction system can be adjusted and a lower annealing temperature can be set accordingly.
3) If the fragment is too long and complex to amplify, has a high probability of mismatch, and has the point mutation, the appropriate annealing temperature (about 60℃), cycle number (about 30 times), and the enzyme amount can be set in the amplification.
2. Enzyme Digestion and Ligation
When encountering double-enzyme digestion during an experiment, problems such as cumbersome enzyme digestion steps, long experimental cycle, star activity, and incomplete enzyme digestion often occur. Therefore, it is key to choose a buffer that can guarantee the activity of the two enzymes. Choosing the right buffer can not only ensure enzyme digestion efficiency, but also helps speed up enzyme digestion time.
3. Transformation and identification
1) Negative and positive controls can be set when low clone numbers are encountered after transformation. During the cloning process, the negative control’s lack of colony growth may be caused by incomplete enzyme digestion. An additional indicator of a cloning issue is the void of spots in the positive control, following transformation.
2) Primers identified via colony PCR should be designed across regions, with one primer on the vector and the other primer on the target fragment. This way, false positive clones can be avoided and reverse insert fragments can be screened out. False clones can be further ruled out by extracting the plasmid, conducting enzymatic digestion, and running the gel.
Plasmid Preparation | Synbio Technologies
Synbio Technologies provides plasmid preparation services from microgram to gram scale for our customers in academia, biotechnology, and biopharmaceuticals fields around the world. We provide both research-grade and transfection-grade plasmids with selectable endotoxin levels (< 100EU/mg, <30EU/mg, <5EU/mg) with zero RNA or bacterial contamination. Our prepared plasmids have stable quality, batch-to-batch consistency, and short cycles. In addition, our plasmid preparation service includes sequence verification, restriction enzyme digestion, and endotoxin analysis. We meet the diverse downstream applications of our customers in various fields, such as transfection, antibody preparation, vaccine development, and gene therapy research.
 DNA Synthesis
DNA Synthesis Vector Selection
Vector Selection Molecular Biology
Molecular Biology Oligo Synthesis
Oligo Synthesis RNA Synthesis
RNA Synthesis Variant Libraries
Variant Libraries CRISPR Libraries
CRISPR Libraries Oligo Pools
Oligo Pools Virus Packaging
Virus Packaging Gene Editing
Gene Editing Protein Expression
Protein Expression Antibody Services
Antibody Services Peptide Services
Peptide Services DNA Data Storage
DNA Data Storage Standard Oligo
Standard Oligo Standard CRISPR Libraries
Standard CRISPR Libraries Standard CRISPR Plasmid
Standard CRISPR Plasmid ProXpress
ProXpress Protein Products
Protein Products
























