A genome-scale metabolic model (GSMM) is a specific microbial metabolic network composed of gene-protein (enzyme) – biochemical reaction associations by integrating genomics, transcriptomes, proteomics, and metabolomics data. GSMMs connect the biochemical reaction information obtained from the genome with the metabolic phenotype and provides an efficient platform for the systematic understanding of microbial physiological metabolic function models.
Applications
- Prediction of Growth Phenotype
- Guidance on Metabolic Engineering
- Analysis of Evolutionary Differences between Species
- Multi-omics Integration Analysis
GSMM Work Flow
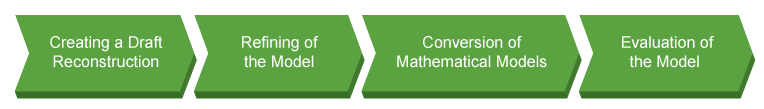
Database and Software
| Number | Database | Number | Software |
|---|---|---|---|
| 1 | Genbank | 1 | COBRA (Matlab) |
| 2 | EMBL | 2 | Simpheny |
| 3 | Ensembl | 3 | MetaFluxNet |
| 4 | Uniprot | 4 | Pathway tools software |
| 5 | MetaCyc | 5 | MosekSolver |
| 6 | Pubchem | 6 | Cytoscape |
| 7 | KEGG | 7 | Adobe Illustrator |
| 8 | Pubmed | 8 | Cell Designer |

