At Synbio Technologies, we are at the forefront of synthetic biology innovation, empowering scientists and researchers to explore the complex world of genetics and biological functions with cutting-edge technology. Our high-throughput sequencing technology has revolutionized the study of transcriptomes, allowing us to comprehensively sequence almost all RNA information in a tissue or organ.
Transcriptomes are critical to understanding the genetic makeup of a particular cell under a specific physiological condition. By analyzing mRNA, rRNA, tRNA, and non-coding RNA, we can gain insights into the expression of genes in cells, tissues, or individuals under different physiological or pathological conditions. This knowledge forms the foundation of genetic research, clinical diagnosis, and drug development.
At Synbio Technologies, we are proud to offer eukaryotic and prokaryotic RNA sequencing services that enable researchers to unlock the full potential of transcriptomes. By harnessing the power of RNA sequencing, we can bond a genome’s genetic information with biological functions, enabling us to develop a deeper understanding of the intricate mechanisms that drive life.
Applications
- In medical research, RNA sequencing has become a powerful tool for understanding the genetic mechanisms behind various diseases. By analyzing the transcriptome, researchers can identify disease markers, diagnose and classify diseases, and develop personalized therapies based on the unique genetic makeup of individual patients. RNA sequencing also plays a critical role in disease recurrence diagnosis and evaluating the clinical efficacy and drug toxicology of treatments.
- In life sciences research, RNA sequencing has numerous applications, including studying abiotic environmental relationships, understanding metabolic pathways and functional genomics, and identifying phenotypic traits in plants and microorganisms. By analyzing the transcriptome of medicinal plants, researchers can discover new therapeutic compounds and better understand the biological mechanisms behind their medicinal properties.
Competitive Advantages
- High Data Quality: Rich experience in library construction for prokaryotic RNA sequencing to reach good rRNA removal efficiency.
- High Coverage: High or low abundances can be identified and quantified simultaneously.
- Strand-specific RNA-seq Library: The dUTP strand-specific RNA-seq library is used to ensure the directivity of transcripts and accurate quantitative results.
- Comprehensive Analysis: Specific probes and reference genomic information are not necessary to detect genes but also to discover new transcripts.
- Integrative Analysis of Multiomics: Full spectrum & comprehensive analysis of biomolecule function and regulatory mechanisms.
Competitive Advantages

Service Specifications
| Service | Sample Type | Sequencing Model | Sampling Requirements |
|---|---|---|---|
| Prokaryotic RNA Sequencing | HiSeq 4000, PE150 | Total RNA ≥3μg, Concentration ≥70 ng/μL | |
| Eukaryotic RNA Sequencing | Cell, tissue, serum, plasma, total RNA, etc. |
Project Design
The design idea of a transcriptome experiment is to compare different genes, and the common type is to compare the experimental group and the control group. With time and space factors considered, multiple comparative analyses can be implemented according to different growth stages or the occurrence and development of diseases. At least 3 biological replicates are required for each group.
Analysis Items
1. Prokaryotic transcriptome sequencing
| Number | Analysis Item | Number | Analysis Item |
|---|---|---|---|
| 1 | Raw data processing and data quality control | 7 | Differential gene cluster analysis |
| 2 | Reference genome alignment | 8 | KEGG enrichment analysis of differential genes |
| 3 | Quality assessment of RNA-Seq | 9 | Antisense transcript prediction |
| 4 | Gene expression level analysis | 10 | Operon analysis |
| 5 | Differential gene expression analysis | 11 | sRNA analysis |
| 6 | GO enrichment analysis of differential genes | 12 | Mutation analysis |
2. Eukaryotic transcriptome sequencing with reference genome
| Number | Analysis Item | Number | Analysis Item |
|---|---|---|---|
| 1 | Raw data output statistics | 7 | KEGG annotation of Unigene |
| 2 | Reference genome comparison and statistics | 8 | GO enrichment analysis of differential genes |
| 3 | Analysis of gene expression abundance | 9 | KEGG enrichment analysis of differential genes |
| 4 | SNP and InDel anslysis | 10 | Prediction of new transcripts |
| 5 | GO enrichment analysis | 11 | Differential splicing analysis |
| 6 | GO annotation for Unigene | 12 | DEU analysis (Differential Exon Usage) |
3. Eukaryotic transcriptome sequencing without reference genome
| Number | Analysis Item | Number | Analysis Item |
|---|---|---|---|
| 1 | Raw data output statistics and quality control | 8 | KOG annotation for Unigene |
| 2 | Transcript splicing | 9 | SNV/SNP analysis |
| 3 | Length distribution statistics and GC content statistics of Unigene and Transcript | 10 | SSR analysis of Unigene |
| 4 | Predict coding protein frame according to splicing sequence | 11 | Analysis of gene expression abundance |
| 5 | Unigene functional annotation | 12 | Differential gene expression analysis |
| 6 | KEGG enrichment analysis | 13 | GO enrichment analysis of differential genes |
| 7 | Profile analysis of differential gene expression | 14 | KEGG enrichment analysis of differential genes |
Data Analysis

Box Diagram of Gene Expression Distribution
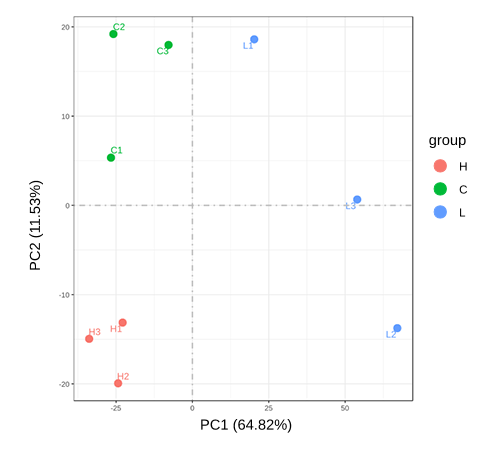
PCA Analysis
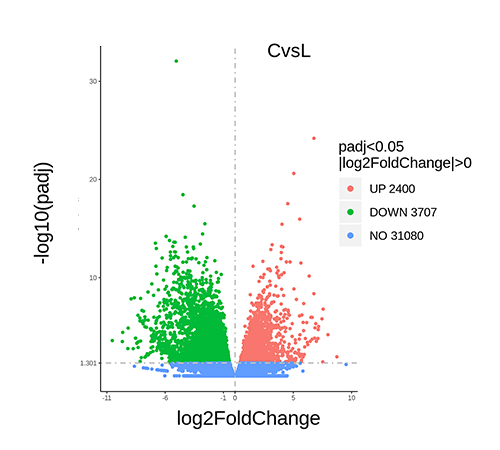
Differential Gene Volcano Plot
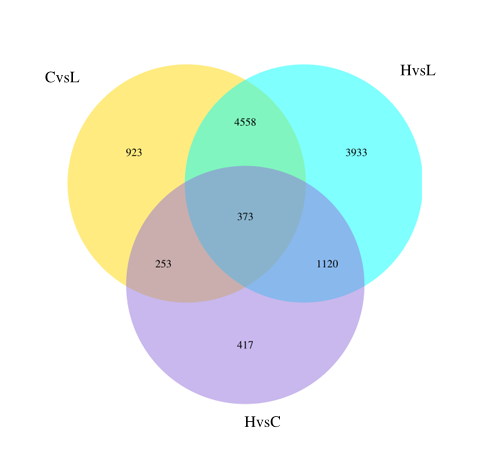
Differential Gene Venn Diagram
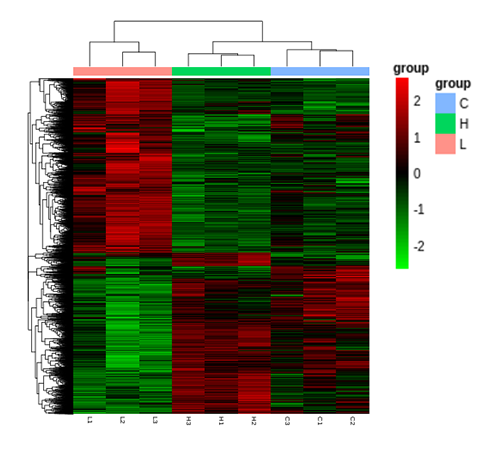
Cluster Heatmap
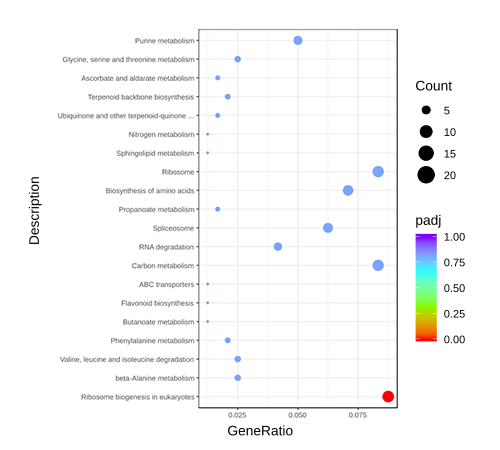
Cluster Heatmap
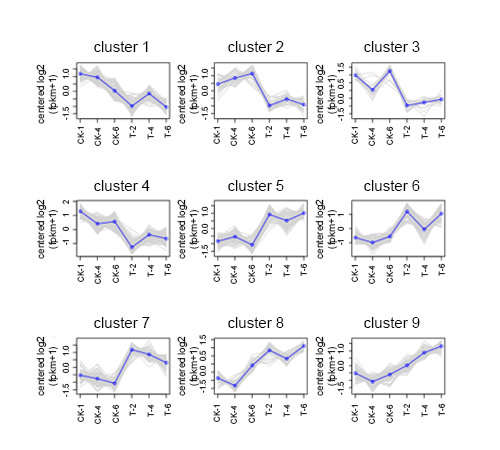
Differential Gene Trend Analysis
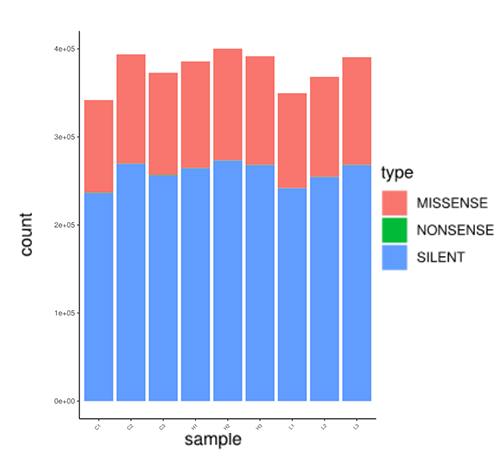
Variation Locus Region Statistics

