Biological evolution is a long, complex process shaped by genetic mutations and natural environmental selection. In today’s rapidly advancing life sciences field, directed evolution has emerged as a crucial tool in mimicking natural evolution. By harnessing the power of directed evolution, researchers can pinpoint variants with improved phenotypes, paving the way for revolutionary advancements in biological function design and enhancement. Mutagenesis libraries serve as treasure troves of diverse mutation sequences, sourced from nature, laboratory experiments, and computational simulations. This innovative approach has proven to be a game-changer in fields like protein directed evolution, antibody discovery, drug target screening, and enzyme optimization.
Mutagenesis Library Types
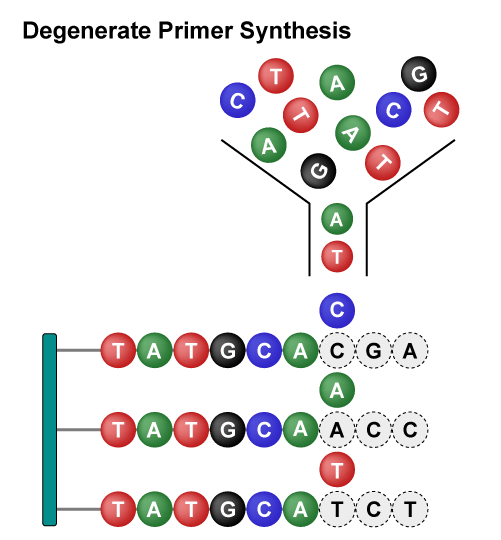
Site-Saturation Mutagenesis Libraries utilize specific degenerate primers (such as NNK), resulting in 32 different codons, including amino acids, stop codons, and synonymous codons. Despite uneven amino acid distribution, this screening method rapidly, and cost-effectively, generates highly diverse mutants that facilitate protein remodeling and research into the relationship between protein structure and function.
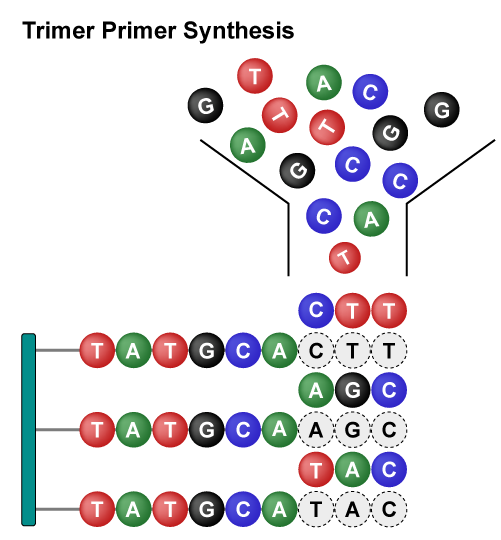
Trimer Combinatorial Mutagenesis Libraries mutate amino acids as a single unit to expand the mutation range. By designing sequences, the types and proportions of amino acid can be flexibly specified, avoiding unnecessary synonymous codons and amino acids. This approach addresses issues such as codon shifting, frameshift mutations, and control over the inclusion of stop codons. Moreover, it significantly enhances library uniformity, coverage, and diversity.
Saturation Scanning Variant Libraries involve the systematic substitution of amino acid residues with alanine. This method provides detailed insights into the role of individual residues in protein functionality, stability, and activity. By creating a comprehensive set of single-residue variants, researchers can pinpoint critical residues and better understand protein structure-function relationships. This approach is particularly useful for mapping functional sites and guiding protein engineering efforts.
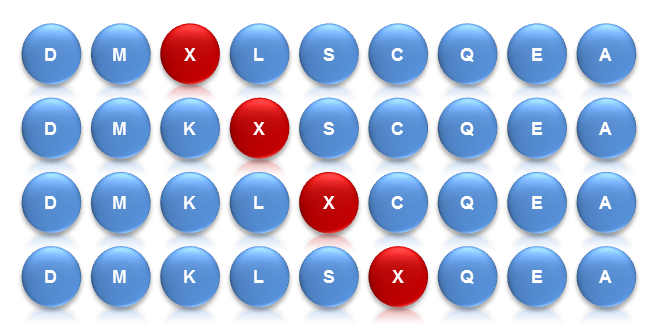
Overall, these libraries employ different strategies to achieve rapid and diversified protein mutations. The Site-Saturation Mutagenesis Library achieves precise mutations of single amino acids, while the Combinatorial Mutagenesis Library achieves precise mutations of multiple amino acids at multiple sites. Saturation Scanning Variant Libraries systematically substitute amino acid residues with alanine to provide detailed insights into the role of individual residues. They each have their own advantages and can be flexibly chosen according to specific research needs, jointly driving the advancement of protein engineering.
Applications of Customized Mutagenesis Libraries
Protein Directed Evolution
- Protein Engineering: By precisely controlling key amino acid mutations in target proteins, we construct protein variants with specific functions or properties, regulating protein activity, stability, and affinity to meet diverse application needs.
- Novel Functions Discovery: Introducing diverse amino acid variations can unveil new protein functions, such as catalyzing new reactions or binding new ligands, thereby facilitating functional screening and evolution of proteins.
- Enzymatic Performance Optimization: Amino acid sequence variations enhance enzyme catalytic efficiency, selectivity, and stability to meet specific catalytic demands under extreme conditions like high temperature or low pH.
- Drug Resistance Research: Simulating drug pressure enables the screening of mutant strains tolerant to drugs, uncovering mechanisms of drug resistance and guiding optimization of drug treatment strategies.
Antibody Development
- Humanized Antibody Design: Introducing specific human amino acids into humanized antibody sequences enhances compatibility and stability, expediting the humanization process.
- Enhanced Affinity: Introducing specific amino acid mutations or variations alters the binding affinity between antibodies and target molecules, improving therapeutic efficacy and selectivity.
- Improved Stability: Structural adjustments of antibodies enhance in vivo stability and half-life, prolonging drug efficacy, reducing dosing frequency, and enhancing patient treatment convenience.
- Reduced Immunogenicity: Avoiding or minimizing sites in antibodies that are antigenic to the human immune system lowers immunogenicity and the risk of adverse reactions, making antibodies more suitable for clinical therapy.
Drug Target Screening
- Target Discovery: Precisely controlling and directing amino acid mutations in specific regions of known targets generates variants with new structural and functional characteristics. These variants may exhibit activities or specificities different from the original targets, providing powerful tools and platforms for discovering and developing new drug targets.
- Target Validation: Adjusting the proportions and combinations of amino acids in mutant strains achieves coverage and testing of various functional properties, comprehensively assessing the activity, affinity, stability, and other critical properties of different mutant strains.
- Drug Screening: High-throughput screening of protein variants with different sequences efficiently identifies targets with high affinity to specific drugs, accelerating the drug screening process and improving drug discovery efficiency.
Synbio Technologies | Drivers of Biological Evolution
We deeply understand that the pursuit of innovative scientific research requires our utmost dedication and comprehensive support. Beyond merely offering services such as trimer primer synthesis and library construction, we prioritize collaboration with you, leveraging our extensive experience and professional expertise to provide guidance throughout your entire research and development pipelines. Our objective is to ensure the smooth implementation of your scientific innovations, leading to the reliable outcomes. Join us and let our variant libraries become the next step to your path to innovation.
When you work with Synbio Technologies, you can effortlessly achieve full coverage of all 20 amino acids, ensuring maximum library diversity. With a large library capacity and industry-leading screening success rate, we confidently propel your scientific research forward. Unlock tailored research solutions and embark on your journey of scientific discovery, today!
References
[1] Prassler, Josef, et al. “HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems.” Journal of molecular biology 413.1 (2011): 261-278.
[2] Chembath, Anupama, et al. “Nondegenerate Saturation Mutagenesis: Library Construction and Analysis via MAX and ProxiMAX Randomization.” Directed Evolution: Methods and Protocols. New York, NY: Springer US, 2022. 19-41.

