Synthetic biology aims to design and build innovative biological components, novel devices, and novel systems, while also redesigning natural biological systems for a myriad of biotechnological applications. As researchers strive to engineer biological systems by introducing new DNA sequences, the demand for tailor-made synthetic DNA sequences have grown considerably. Gene synthesis typically encompasses fragments of less than 10kb, serving small-scale biological modifications, while research applications at the genome level often involve gene sequences spanning 10-500kb in length. However, when designing complex genomes, it is essential to synthesize genes at the mb level.
| Type | Principle | Specificities | Limitations |
|---|---|---|---|
| Column-Based Synthesis | Phosphoramidite-based Synthesis of Oligonucleotides | Mature, Automated Production Equipment Wide Range of Applications Low Error Rate |
Low Throughput Synthesis Length up to 200nt |
| DNA Chip Technology |
Inkjet Microarray | High-Throughput Low Cost Long Oligos |
Synthesis Length from 20-300nt Synthesis Accuracy Lower than Column-Based Synthesis Inconvenient for Assembly and Splicing |
| Photochemistry Method | High-Throughput Low Cost Short Oligos |
||
| Electrochemical Method | High-Throughput Low Cost Long Oligos |
||
| Semiconductors Combined with Electrochemistry | Ultra High-Throughput DNA Synthesis Low Cost Long Oligos |
Patent Limitations | |
| Enzymatic Synthesis | Microarray Method Yeast in vivo DNA Synthesis Enzymatic DNA Synthesis |
Fast Synthesis Eco-Friendly Low DNA Mutation Rate |
Technology Immaturity Early Commercialization Stage |
DNA Assembly Technology
Overlap extension PCR (OE-PCR), BioBricks™, Golden Gate, TPA (Twin-primer non-enzymatic), Gibson, and TAR yeast homologous recombination technologies are commonly used DNA assembly techniques. Currently, Gibson assembly is the most popular way to achieve directed cloning and in vitro multiple-segment assembly. It can connect multiple DNA segments automatically without DNA polymerases and ligases, making it possible to construct a plasmid over 10kb with fast turnaround.
| DNA Assembly | Gibson Assembly | Yeast Homologous Recombination |
|---|---|---|
| DNA Polymerases / Taq Ligases | Required | Not Required |
| Multiple Fragment Assembly | Seamless Assembly | One-Step Assembly |
| Long DNA Fragments and Genomes | Difficulty Synthesizing Fragments over 20kb | Able to Assemble Fragments up to 1.08mb |
What Affects the Accuracy of DNA Synthesis and Assembly?
The main factors affecting the accuracy of DNA synthesis and assembly includes initial oligo synthesis efficiency and the number of DNA splices. While the chemical synthesis method has achieved an impressive 99.8% efficiency in single-step oligo synthesis, we still need to overcome challenges posed by polymerization sequences, long repeat sequences, and abnormal DNA structures that inhibit successful DNA assembly. Additionally, it is crucial to consider that the more assemblies required to achieve the desired DNA, the higher the associated costs for quality control measures such as clone selection and sequencing.
The synthetic assembly of DNA longer than 10kb is one of the major technology development challenges in the field of gene synthesis. Reducing synthesis errors and costs is the goal for technological advancement in this field. Optimization of complex sequences by computer algorithms and development of intelligent assembly techniques will help to solve the problem of assembling complex and long DNA in an efficient, cost-effective manner.
Experience the Revolution of DNA Synthesis and Assembly at Synbio Technologies
Synbio Technologies has advanced DNA synthesis and assembly technologies, providing customized solutions to researchers around the globe for long and complex gene synthesis. Our bio-intelligent analysis tools, Complexity Index (CI), NG Codon, and AI-TAT, paired with our proprietary reagents, Syno Interlinkage and Syno Assembly, propel DNA synthesis forward with enhanced intelligence, precision, and speed.
- Complexity Index (CI) Makes Gene Synthesis Easier
Our CI (Complexity Index) system can intelligently analyze DNA for complex genes with repetitive sequences, hairpin structures, high GC content, low GC content, poly structures, and more. This proprietary technology enables us to efficiently optimize DNA synthesis and assembly strategies.
- AI – Turnaround Time Makes Gene Synthesis Efficient
At Synbio Technologies, we pride ourselves on our proprietary AI-TAT production cycle prediction system. This proprietary technology allows us to accurately predict production cycles that are incredibly close to the actual ones. By combining scientific prediction and efficient delivery, we empower our customers to effectively plan and rationalize the progress of their experiments.
- NG Codon Makes Gene Synthesis More Refined
The 2.0 upgrade of our NG Codon optimization system is a sophisticated solution designed to enhance protein expression. This advanced version considers a multitude of factors, including codon preference, mRNA secondary structures, GC content, hairpin structures, translation initiation, and termination efficiency. The result? A more refined and efficient codon optimization design.
Our monthly synthesis throughput reaches tens of thousands of sequences, capable of synthesizing single DNA strands up to 150kb in length. Whether it’s a simple DNA fragment or a complex and long pathway or genome, we boast an impressive 100% success rate!
- Fragment XP | DNA and Gene Fragments
Our Fragment XP gene fragments are synthesized at 125-3000bp in length and shipped in as fast as 3 days! We deliver lyophilized PCR products that save you time, effort, and cost for applications in genetic engineering, gene editing, protein expression and function research, antibody drug screening, vaccine research, and many other fields. - Gene Synthesis
At Synbio Technologies, we offer you the ultimate solution for gene synthesis – fast, high-quality, and efficient services. With our expertise, we guarantee 100% accuracy in delivering precisely synthesized DNA that can be easily cloned into your desired vector. - Small Genome Synthesis
Synbio Technologies provides one-stop services for long gene synthesis and assembly as well as gene cluster / small genome synthesis and assembly. We guarantee 100% accuracy of your DNA products.
Case Study
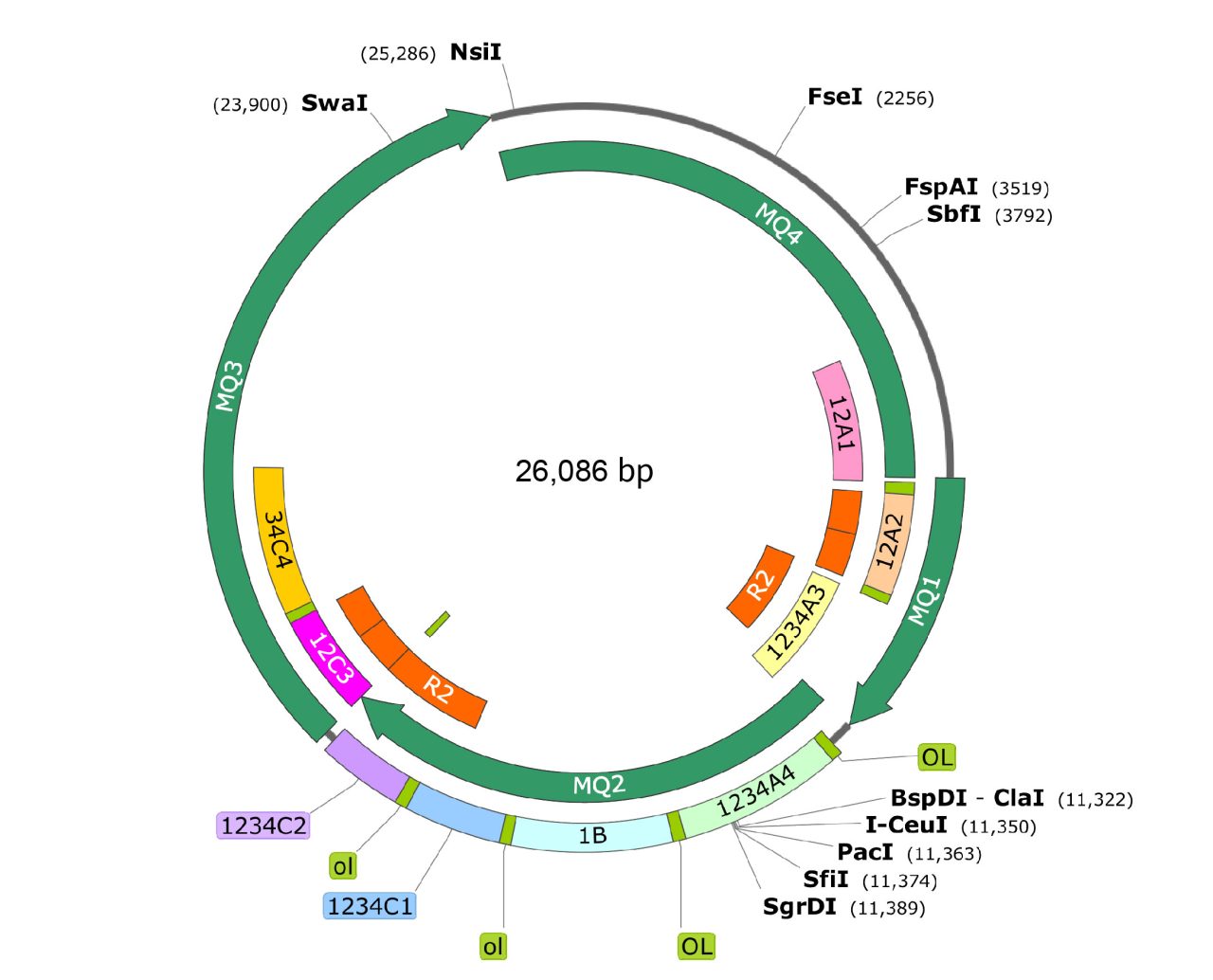
14kb Complex DNA Synthesis and Assembly
We synthesized a 14kb complex sequence and assembled it into our in-house pSynoYac0-Cm vector.
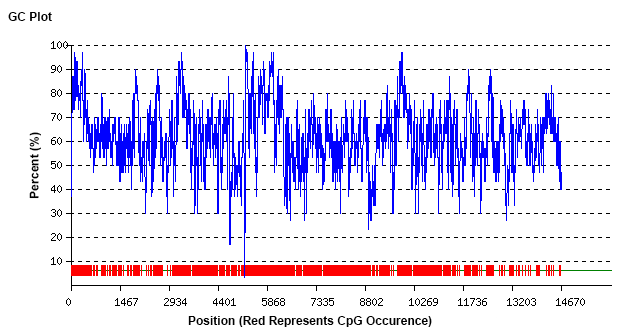
| Sequence Complexity | Extreme |
|---|---|
| Sequence Length | 14,675bp |
| GC Content | 61.9 | Normal |
| GC 50nt Volatility | 72 | Complex |
| GC 50nt Content | 98-26 | Complex |
| Base Repeatability | A7C10G16T6 | Complex |
| Number of Long Repeats (20nt) | 26 | Extreme |
| Number of Short Repeats (9nt) | 18.4% | Normal |
| Palindromic Sequence Length | 16 | Normal |
| Reverse Repetition Length | 20 | Extreme |
Complexity Index (CI) Analysis
- The GC content of the target DNA fragment fluctuated greatly, and the local region was even as high as 100%.
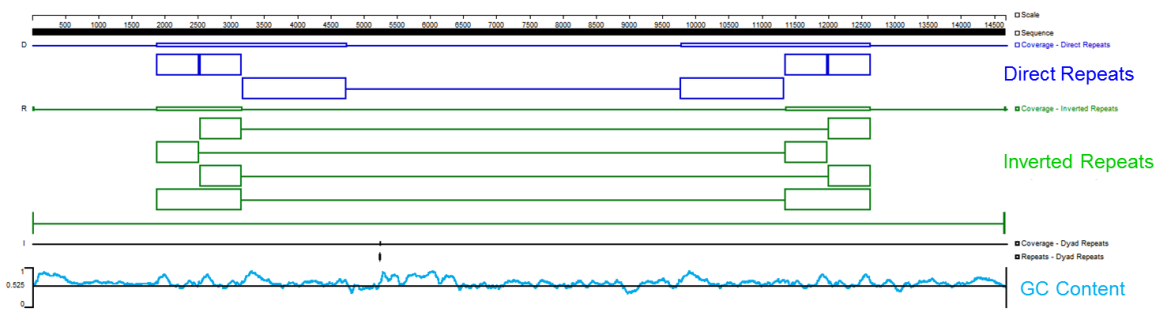
- The target DNA fragments have multiple repetitive fragments, including 4 direct repeats of 649bp and two of 1579bp fragments, as well as four reverse repeats of 649bp, and two of 1299bp fragments.
Solutions
- In this complex GC content scenario, we adjusted the polymerase content, ions, and other components in the PCR reaction system. Additionally, we have optimized critical parameters within the PCR amplification program, such as annealing temperature and extension time. These important adjustments have been made to guarantee both the success rate and fidelity of PCR chain assembly.
- In this repetitive sequence scenario, we have carried out targeted DNA codon design and optimization by NG Codon, while utilizing enzymatic ligation, Golden Gate, and Gibson assembly to assemble the intermediate construction of synthetic repetitive sequences.
To ensure the utmost accuracy, we employed advanced NGS technology to validate the sequencing of full-length genes. Additionally, we utilized Sanger sequencing to confirm the accuracy of repetitive sequences. The results of these two methods align seamlessly, providing a comprehensive validation of our findings. Rest assured, the final constructs we deliver are 100% compatible with the precise target sequences required by our customers.
References
Jiang XE, Wang Y, Shen Y. The review of DNA synthesis technologies and instruments development [J]. Journal of Integration Technology, 2021, 10(5): 80-95.
Chang H, Wang C, Wang P, Zhou J, Li B. DNA assembly technologies: a review. Sheng Wu Gong Cheng Xue Bao. 2019 Dec 25;35(12):2215-2226.
Hughes RA, Ellington AD. Synthetic DNA Synthesis and Assembly: Putting the Synthetic in Synthetic Biology. Cold Spring Harb Perspect Biol. 2017 Jan 3;9(1):a023812.
Ma S, Saaem I, Tian J. Error correction in gene synthesis technology. Trends Biotechnol. 2012 Mar;30(3):147-54. doi: 10.1016/j.tibtech.2011.10.002.
Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009 Feb;37(2):e16. doi: 10.1093/nar/gkn991.
Chang H, Wang C, Wang P, Zhou J, Li B. DNA assembly technologies: a review. Sheng Wu Gong Cheng Xue Bao. 2019 Dec 25;35(12):2215-2226.