Respiratory viruses are capable of causing a wide range of respiratory infections from minor colds to severe illness. Timely and accurate detection of these viruses is essential for optimized patient care, infection control, and public health interventions. But how can we improve the accuracy and efficiency of virus detection? That’s where PROBES come in!
Explore the World of RT-PCR
Reverse transcription polymerase chain reaction (RT-PCR) has become a powerful tool for the diagnosis of respiratory viruses due to its high sensitivity and specificity. This is a method of nucleic acid quantification that incorporates a fluorescent probe into the RT-PCR reaction system. This technique relies on the use of specific probes that emit fluorescence during amplification.
- Sensitivity and Specificity: RT-PCR probes demonstrate high sensitivity and specificity in detecting respiratory viruses, enabling the differentiation between different viral strains.
- Multiplexing: The use of multiplex RT-PCR probes allows simultaneous detection of multiple respiratory viruses in a single reaction, providing a comprehensive diagnostic approach.
- Quantification: RT-PCR probes facilitate the quantification of viral load, aiding in assessing the severity of infection and monitoring the progress of treatment.
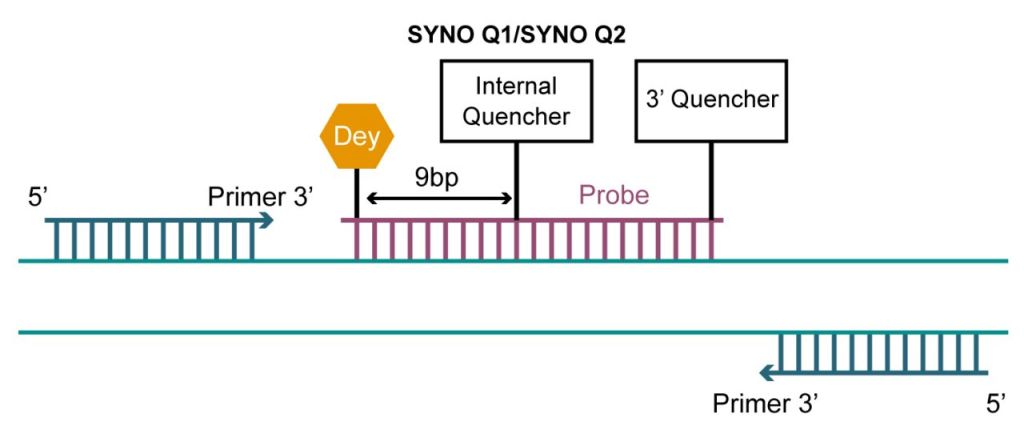
Double-Quencher Probes
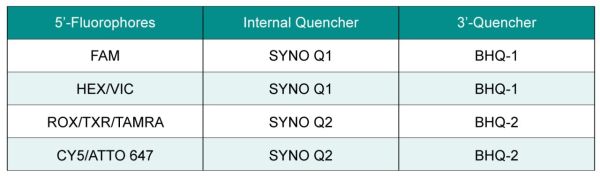
Conventional single-quenching probes have a fluorescent group at the 5′ end and a quenching group at the 3′ end, while double-quenching probes add an additional quenching group inside the probe to optimize the quenching effect. Double-quencher probes employ two mechanisms of fluorescence quenching to enhance sensitivity and reduce background fluorescence. Additional quenching groups added in the middle of the probe are the SYNO Q1 or SYNO Q2 quencher.
The combination of different quenching mechanisms increases the efficiency of fluorescence suppression and reduces background fluorescence. This reduction in background signal enhances the signal-to-noise ratio, improving the accuracy and reliability of the assay. While conventional probes are typically 20-30 bp in length, double-quencher probes can be designed to be much longer, up to 40 bases. Furthermore, this design enhances the stability of the double-stranded structure and prevents the enzymatic action of nucleic acid exonucleases, which can be applied to in situ hybridization of anti-miRNAs as well as miRNAs and mRNAs.
Double-quencher probes represent an advanced design strategy to achieve highly sensitive and specific detection in various molecular and cellular applications. Their versatility makes them valuable tools in both research and diagnostics.
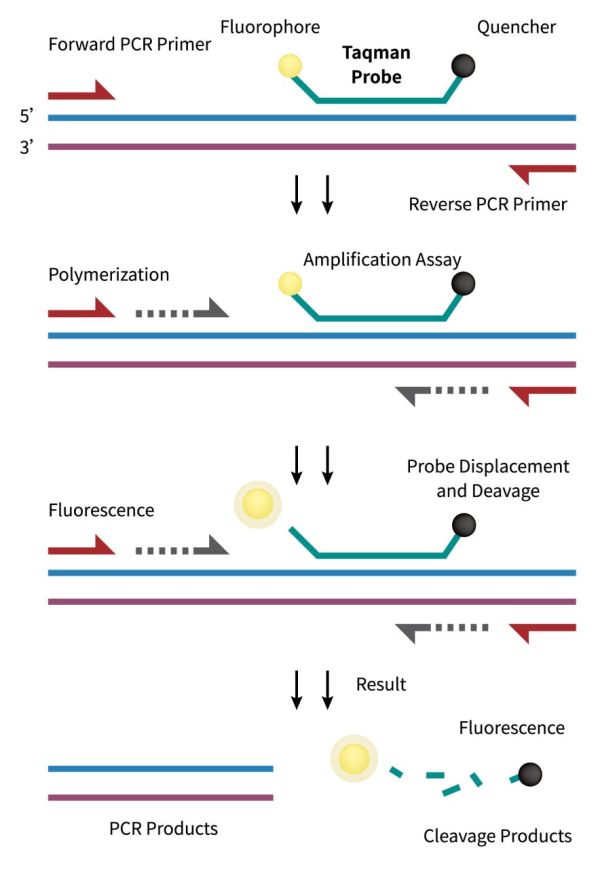
TaqMan Probes
TaqMan probes contain a fluorescent group at the 5′ end and a quenching group at the 3′ end. The probe is designed to ensure that the probe hybridizes only to the desired target sequence within the PCR amplicon. It takes advantage of the 5′ to 3′ exonuclease activity of the Taq polymerase. During the extension phase of PCR, the polymerase cleaves the probe, resulting in the separation of the fluorescent reporter dye from the quencher, resulting in a measurable increase in fluorescence. The real-time fluorescence signal allows researchers to track the amplification kinetics continuously.
TaqMan probes contribute to the reduction of false-positive signals by providing an additional layer of specificity. It enables quantitative analysis of the initial amount of target nucleic acid in the sample. The fluorescence signal generated during each PCR cycle is directly proportional to the amount of amplified DNA. This quantitative aspect makes TaqMan probes particularly valuable in applications such as gene expression analysis and viral load quantification. Furthermore, TaqMan probes are compatible with multiplex PCR reactions, allowing the simultaneous detection and quantification of multiple target sequences in a single reaction. This capability is advantageous for studies requiring the analysis of multiple genes or the identification of diverse pathogens within a complex sample.
TaqMan probes are widely used in diagnostic applications for the detection of infectious agents, genetic mutations, and other nucleic acid targets. Their high specificity and real-time monitoring capabilities make them valuable tools for rapid and accurate diagnostic assays.
Molecular Beacon Probes
Molecular Beacons are hairpin-shaped oligonucleotide probes. The 5′ end of the molecular beacon is labeled with a fluorophore (e.g., FAM, HEX), and the 3′ end is labeled with a quencher (e.g., DABCYL, BHQ). In the absence of a target, the proximity of the quencher suppresses the fluorescence of the fluorophore. When the probe binds to the target sequence, the hairpin opens, separating the fluorophore and quencher and resulting in fluorescence emission. The stem-loop structure of molecular beacons keeps them in a hairpin conformation when unbound. This conformation prevents the interaction of the fluorophore with the surrounding environment, minimizing background fluorescence.
Molecular beacons offer high specificity and enable the quantification of viral RNA or DNA in a sample by providing real-time fluorescence signals proportional to the amount of amplified nucleic acid during PCR. Molecular beacons can be employed for genotyping and mutation analysis of viral genomes. The sequence specificity of the beacon allows discrimination between different viral strains or the identification of specific mutations.
The principles of molecular beacon probes, with their stem-loop structure and real-time monitoring capabilities, make them versatile tools for the detection and analysis of viruses, contributing to various applications in molecular diagnostics and research.
Revolutionize Your Virus Detection at Synbio Technologies
Unlock the potential of RT-PCR probes and take your research to new heights with Synbio Technologies! Between our wealth of chemical modification groups, fluorescence, and quenchers and nearly 20 years of oligonucleotide synthesis experience and powerful modification abilities, we can provide various diagnostic probe products for use in molecular hybridization and PCR, such as: TaqMan/MGB probes, molecular beacons, double-quencher Probes, LNA Probes, and more.
Why Choose Us?
- A Probe for Every Purpose: Whether it’s TaqMan/MGB, Molecular Beacons, or LNA Probes, we’ve got you covered. And that’s just the beginning!
- Purity is Our Promise: Thanks to top-notch purification technology, our probes boast impressive purity. Say goodbye to pesky background noise!
- Quality? Checked! With meticulous removal of small molecule fluorescent dye residues and a 100% LC-MS quality check for each probe, rest easy knowing quality is our middle name.
- Tailored Just for You: Need a specific amount or purification method? From nanomoles to micromoles, HPLC to PAGE, we personalize to your precise specifications, maintaining high purity and standards all the way.
References
- Schöllkopf S, Knoll A, Homer A, Seitz O. Double FIT hybridization probes – towards enhancing brightness, turn-on and specificity of RNA detection. Chem Sci. 2023 Mar 23;14(15):4166-4173. doi: 10.1039/d3sc00363a.
- Hirotsu Y, Mochizuki H, Omata M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. J Virol Methods. 2020 Oct;284:113926. doi: 10.1016/j.jviromet.
- Socher E, Knoll A, Seitz O. Dual fluorophore PNA FIT-probes–extremely responsive and bright hybridization probes for the sensitive detection of DNA and RNA. Org Biomol Chem. 2012 Sep 28;10(36):7363-71. doi: 10.1039/c2ob25925g.
- Cheng H, Liu Y, Fu M, Liu J, Chen G. Taqman-MGB FQ-PCR for Human GPIIIa, GP1BA and PEAR1 SNPs. Clin Lab. 2022 Oct 1;68(10). doi: 10.7754/Clin.
- Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR 3rd, Smith TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006 Jan;19(1):165-256. doi: 10.1128/CMR.19.1.165-256.

