mRNA vaccines are the third generation of nucleic acid vaccine developed on the basis of the first generation of inactivated vaccines or attenuated live vaccines and the second generation of subunit vaccines. The active component of mRNA vaccines is RNA, which has two forms. One form is non-replicating mRNA, composed of the antigen encoding sequence and two sides of 5’UTR and 3’UTR. The other form is self-replicating mRNA, where the antigen encoding sequence is constructed in the genome of RNA virus and it can be self-amplified with the help of the virus encoding replicase. Ultimately, this improves the expression level of the antigen gene in the cell under the condition of very low injection dose.
Why do mRNA vaccines stand out?
mRNA vaccines have many unique advantages, which is the main reason to promote its extensive research and application. Easy preparation is one of the main reasons why mRNA vaccines stand out. Its active RNA component is simple, and it can be obtained by in vitro transcription using linear DNA as a template. In this process, the stability and translation efficiency of mRNA can be improved through codon optimization, nucleoside modification, and an auxiliary delivery system, which ensures high specificity and strong stability of the nucleic acid vaccine. In addition, the safety of the vaccine is also a major factor in promoting its widespread use. The mRNA does not integrate into the host genome, so there is no potential risk of infection or insertion mutation and it can be degraded through normal cellular processes.
Technology as a Safeguard for In Vitro Transcription
NGTM Codon optimization technology makes codon optimization more convenient.
The secondary structure of mRNA affects the expression and stability of mRNA in cells. Synbio Technologies independently developed our NGTM Codon optimization technology, which can effectively optimize mRNA sequences and improve mRNA translation efficiency by optimizing codon composition, adjusting GC content, simplifying the secondary structure of mRNA, and removing modules that are not conducive to efficient expression.
Using a vector containing a poly(A)tail more than 100bp and synthesizing with a 100% accurate DNA template sequence.
The template preparation methods for in vitro transcription include PCR and plasmid linearization. During the linearization of plasmids, flat-terminal enzymes or class II restriction endonuclease enzymes (eg.BspQI) are usually used to produce blunt or 5 ‘end double-stranded DNA templates. Synbio Technologies has created the vector containing the poly(A) tail more than 100bp, providing technical support for template preparation in a fast, efficient method.
In Vitro Transcription of RNA
Synbio Technologies uses in vitro transcription technology to synthesize various types of RNA such as sgRNA, lncRNA, and mRNA. Our platform uses a linear DNA sequence as the template, while using T7, T3, or SP6 RNA polymerase to synthesize RNA sequence from DNA.
Add Cap, Tail, and Nucleoside Modifications
A mature mRNA is composed of cap structure, 5 ‘UTR, 3′ UTR, an open reading frame (ORF) and a poly(A) tail. The cap structure and poly(A) tail form a “closed loop”, which is closely related to mRNA stability and translation efficiency. Synbio Technologies can add different modifications to RNA sequences according to customers’ needs to further improve the stability and translation efficiency of RNA products.
Cap Structure
In eukaryotes, the traditional cap structure has three types: m7GpppXpYp (Cap0), m7GpppXmpYp (Cap1), and m7GpppXmpYmp(Cap2). When the 7th carbon atom of G is methylated to form m7GpppN, the Cap is called “Cap 0”; Based on Cap0, if the 2´-O site of the first nucleotide of the transcript is also methylated, it is called “Cap 1”; If the 2´-O sites of both the first and second nucleotides of the transcript are methylated, it is called “Cap 2”. Usually, the cap is added in vitro by adding vaccinia virus cap enzymes or cap analogizing. Cap1 structure is widely found in mRNA of eukaryotes. Studies have shown that mRNA with Cap1 structure modified by in vitro transcription has higher stability and translation efficiency.
Poly(A) Tail
Most mRNA degradation starts from the Poly(A) tail. The Poly(A) tail of mRNA will be reduced to an oligo A tail structure by deadenylation, so the Poly(A) tail is crucial to resist the degradation of mRNA. According to our experimental results, the stability and translation efficiency of mRNA containing a 120nt Poly(A) tail were higher than those containing a 40nt or 80nt Poly(A) tail length.
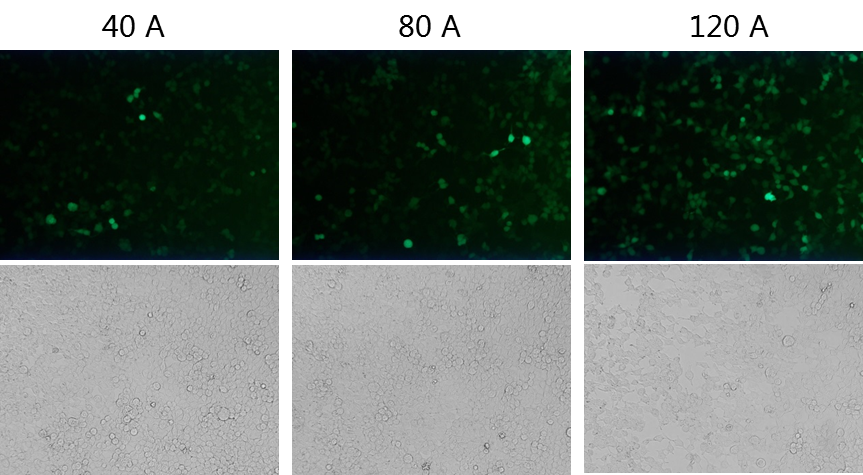
Case study: The effects of different ploy(A) tail on transcription
Modified Nucleosides
In mammals, common mRNA modifications include N1- and N6-methyladenosine (m1A, m6A, m6Am), 3- and 5-methylcytosine (m3C, m5C), 5-hydroxymethylcytosine (hm5C), pseudouridine (ψ) and 2′ -0-methylation (Nm). Studies founded that mRNAs can activate the cellular innate immune system by stimulating toll-like receptor TLRs (TLR3, TLR7, TLR8). However, most of the TLRs are no longer activated with the synthesized sequence using nucleoside analogues [2], such as pseudouridine (ψ), 5-methylcytidine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), and 2-thiuridine (s2U). Therefore, in the development of mRNA vaccines, modified nucleosides are usually introduced into the sequence to improve the stability of mRNA sequence and reduce its immunogenicity.
Synbio Technologies, with leading bio-design and technology, constantly improves our own synthesis abilities to meet the market demand and the comprehensive scientific research needs of our customers.
Related Services
NG™ Codon Optimization Technology
Gene Synthesis
In Vitro Transcription
References
[1] Pardi, N., Hogan, M., Porter, F. et al. mRNA vaccines — a new era in vaccinology. Nat Rev Drug Discov 17, 261–279 (2018).
[2] KORMANN MS, HASENPUSCH G, ANEJA MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice [J]. Nat Biotechnol, 2011, 29(2): 154-157.
[3] Yasmin Granot, Dan Peer,Delivering the right message: Challenges and opportunities in lipid nanoparticles-mediated modified mRNA therapeutics—An innate immune system standpoint, Seminars in Immunology 34, 68-77(2017).

