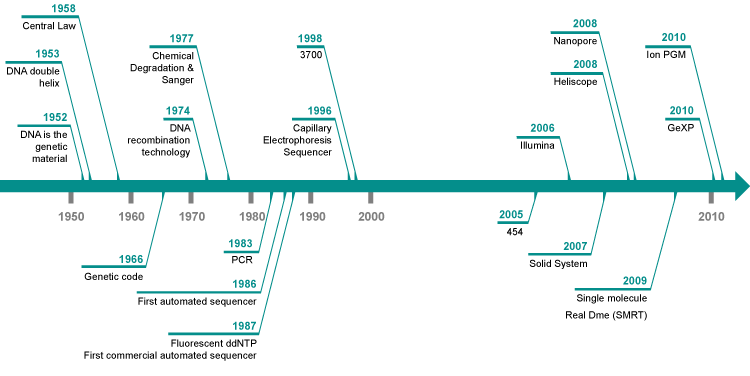
Ever since Watson and Crick unveiled the DNA double helix structure in 1953, the field of sequencing technology has witnessed steadfast and remarkable advancements.
In 1977, British biochemist Laureate Frederick Sanger invented the double-deoxygenated strand termination method of nucleic acid sequencing, which has since been considered the gold standard for nucleic acid sequencing. Based on the name of the inventor, this method is also known as Sanger sequencing. The first generation of DNA sequencing technology, represented by Sanger sequencing, supported the completion of the Human Genome Project and greatly promoted understanding of human being’s own genetic coding information.
With the completion of the Human Genome Project, people entered the era of functional genomes, and traditional sequencing methods could no longer meet the needs of large-scale genome sequencing such as deep sequencing and repeated sequencing, which prompted the birth of Next-Generation Sequencing (NGS) technology. NGS sequencing technology is most notable for its high throughput, sequencing hundreds of thousands to millions of DNA molecules at a time, dramatically increasing sequencing speed while greatly reducing sequencing costs and maintaining high accuracy, but the sequence reading length is generally low. NGS sequencing has been widely used in genomics and has many applications in various fields of research, medicine and industry. The following are some common applications of NGS sequencing:
- Genomic Research: NGS enables the comprehensive analysis of an organism’s entire genome, facilitating the study of genetic variation, gene expression, and genome structure. Researchers can investigate the genetic basis of diseases, identify disease-associated genes, and understand the complexity of human genetic variation.
- Cancer Genomics: NGS helps in understanding the genetic mutations and alterations that drive the development and progression of cancer. It can be used to identify specific mutations, assess tumor heterogeneity, and guide personalized treatment options, such as targeted therapies and immunotherapies.
- Microbial Genomics: NGS allows the sequencing and characterization of microbial genomes, aiding in the identification of pathogens, understanding microbial diversity, and investigating the role of microbes in various environments, including the human microbiome.
- Clinical Diagnostics: NGS has transformative potential in clinical diagnostics. It enables the identification of disease-causing mutations, genetic predispositions, tumors and rare genetic disorders, providing fundamental data for precision medicine. NGS-based tests can be used for prenatal screening, carrier testing, and diagnosis of genetic diseases.
- Infectious Disease Surveillance: NGS can track and monitor the spread of infectious diseases, such as viral outbreaks or antimicrobial resistance. It enables rapid identification and characterization of pathogens, facilitating effective public health responses.
- Agriculture and Food Safety: NGS can be used to enhance crop breeding programs by identifying genes associated with desirable traits, improving plant breeding efficiency, and ensuring food safety by detecting contaminants and pathogens in food products.
- Forensic Analysis: NGS has applications in forensic genetics for human identification, paternity testing, and criminal investigations. It can provide highly sensitive and accurate DNA profiling from minute samples.
These are just a few examples of the broad range of applications for NGS sequencing. The technology continues to evolve, and its increasing speed, accuracy, and cost-effectiveness open up new possibilities in various fields.
Synbio Technologies︱NGS sequencing service
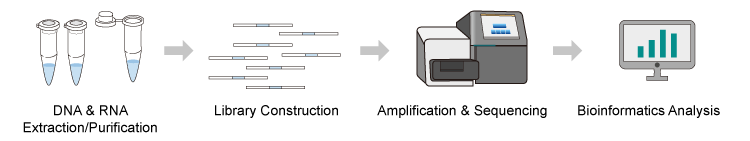
In the process of exploring synthetic biology, Synbio Technologies has accumulated very profound experience in the field of DNA sequencing. We have established the Sanger double deoxy termination analysis platform, which is suitable for sequencing common cloning as well as synthetic projects, and screening targets by next-generation sequencing technology (NGS) for engineering or large-scale screening projects. Our experienced engineers provide stable and reliable gene sequencing and bioinformatics analysis services to researchers around the world using our advanced laboratory equipment.
Sequencing Services
Genome Sequencing
Genomic DNA sequencing enables researchers to obtain sequence information for parts of the genome and to discover new genomic sequences from organisms of any origin. We offer a variety of related services, including library preparation, sequencing, and data analysis for samples derived from a variety of organisms and tissue types. Practical applications of these experiments include genetic variation analysis and the discovery of new genes through de novo assembly.
Transcriptome sequencing
We provide rapid, high-quality transcriptome sequencing for a wide range of applications including gene expression mapping, SNP detection, variable splicing, and transcript annotation. We have developed specific library preparation methods that meet the requirements of high specificity and low sample input.
sRNA sequencing
sRNA sequencing can detect miRNA, siRNA and piRNA simultaneously. Among them, miRNA analysis has a wide range of applications in the discovery of new biomarker miRNA expression maps and the search for miRNA target genes. We provide sRNA library preparation, sequencing and data analysis services for total RNA samples from various organisms and tissue types.
PCR Amplification Sequencing
Amplification sequencing is the sequencing of PCR products with higher reading depth and coverage. It allows not only accurate mutation screening, but also precise detection of low frequency mutations. Amplification sequencing can complete sequence determination of hundreds of PCR amplicons from any source at once.
16S/18S/ITS amplicon sequencing
Amplicon sequencing products mainly include 16S, 18S and ITS region amplification sequencing, which can study microbial diversity of various samples such as environment and intestine. Bacterial diversity is sequenced by 16S amplification, and fungal diversity is sequenced by ITS amplification. Simultaneous studies of bacterial and fungal diversity in an environment can be performed on one sample by both 16S and ITS amplification sequencing.
References
[1] Levy S E , Myers R M .Advancements in Next-Generation Sequencing[J].Annu Rev Genomics Hum Genet, 2016, 17(1):95-115.DOI:10.1146/annurev-genom-083115-022413.
[2] Hu T , Chitnis N , Monos D ,et al.Next-generation sequencing technologies: An overview[J].Human Immunology, 2021.DOI:10.1016/j.humimm.2021.02.012.
[3] Ayman,Grada,Kate,et al.Next-Generation Sequencing: Methodology and Application[J].Journal of Investigative Dermatology, 2013.DOI:10.1038/jid.2013.248.