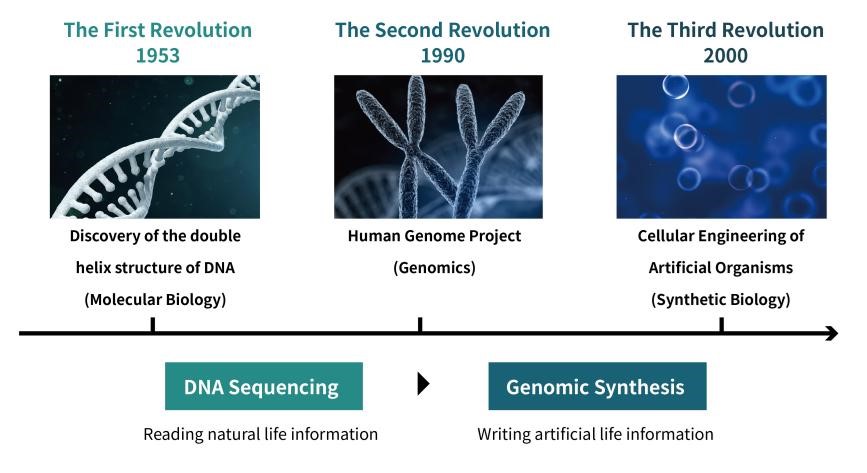
Synthetic biology is a groundbreaking revolution in biotechnology, where engineering methods are used to craft and create organisms with distinctive characteristics. This cutting-edge system leverages the power of enzyme engineering, gene synthesis, sequencing and editing to unlock revolutionary possibilities for biological design.
Synthetic biology has opened the door to unprecedented opportunities for enhancing life activities at an astounding level. By leveraging a method known as ‘Design-Build-Test-Learn’, we are now able to create complex DNA sequences and entire metabolic systems from nucleotide down – leading us closer than ever before towards building entirely new forms of functional, living organisms.
Genome Synthesis
Synthetic biology has made vital steps forward with the advent of genome synthesis. Starting from Sanger Sequencing to Next-Generation and Third-generation sequencing technologies, we are continuously discovering deeper levels of insight into genes, genomes and molecular structures that have enabled us to begin “writing” our own biological codes. Our understanding on basic life forms is now at its greatest ever level due in no small part to these advancements in genomics.
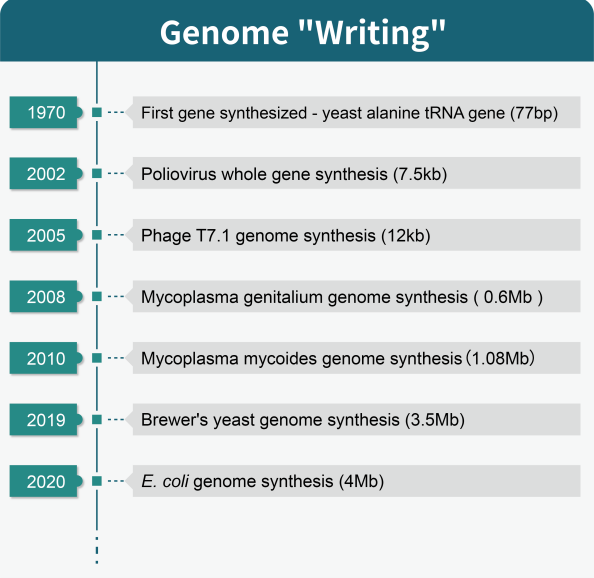
Over the past 50 years, advances in gene synthesis have enabled scientists to progress from a single patented yeast alanine tRNA gene of 77bp up to an incredible 4Mb E. coli genome – setting new records all along the way. We’ve seen success with existing and unnatural genes, prokaryotes and eukaryotes alike within Kb-Gb ranges, leading us ever closer toward attempting complex mammal genomes or even our own natural human DNA – though commercial products still rarely exceed 10kb due largely in part to difficulties surrounding large fragment delivery. With so much potential on the horizon for synthetic genomics research capabilities necessarily requiring efficient transport of greater volumes than previously achieved, it’s clear that mastering this feat is key towards unlocking further unprecedented successes. Leading technologies in oligo and gene synthesis are revolutionizing the industry. With incredible gains being made to reduce costs, boost throughput rates, and expand DNA length possibilities; these advances open up countless opportunities.
DNA Synthesis and Assembly Solutions
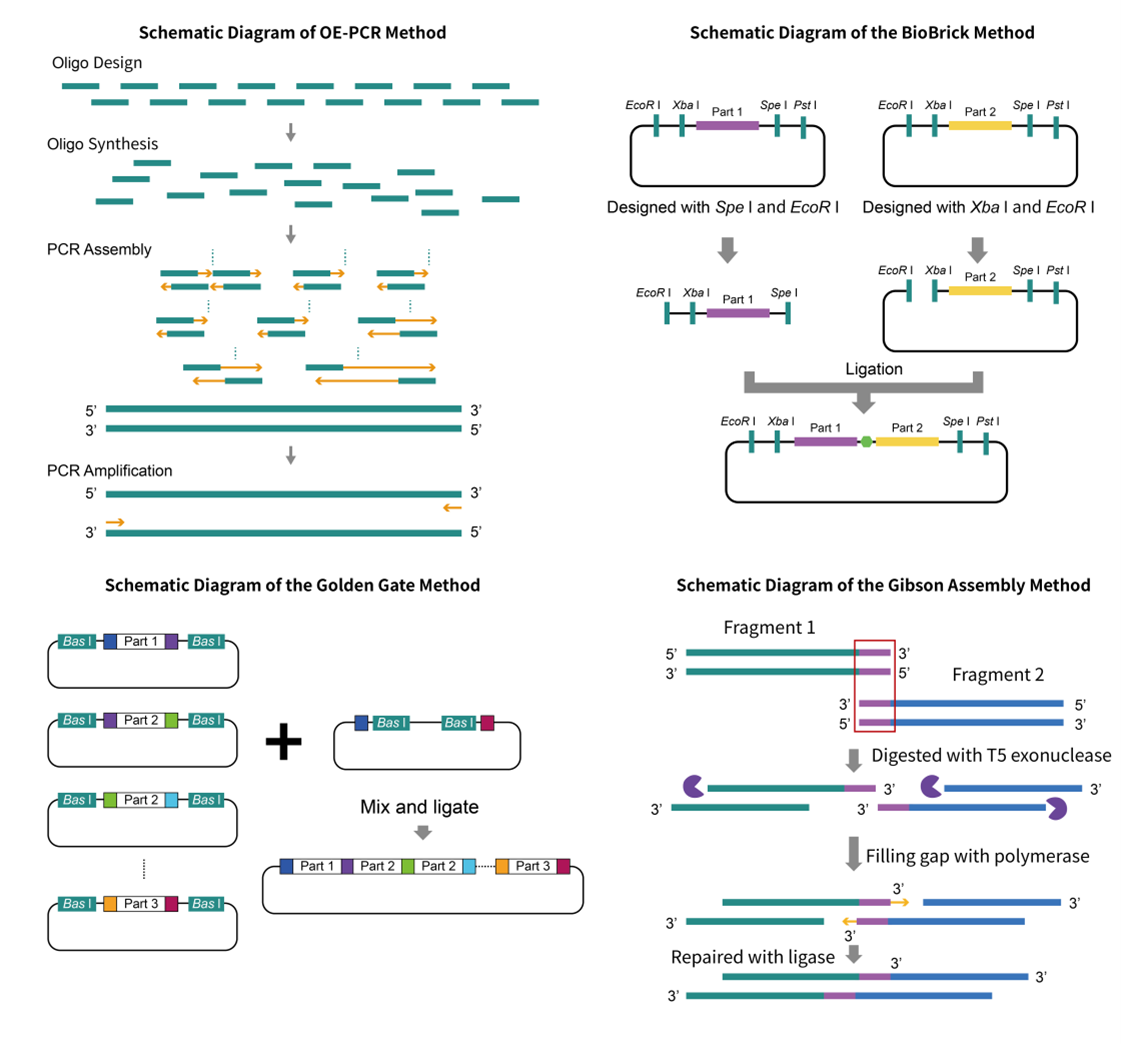
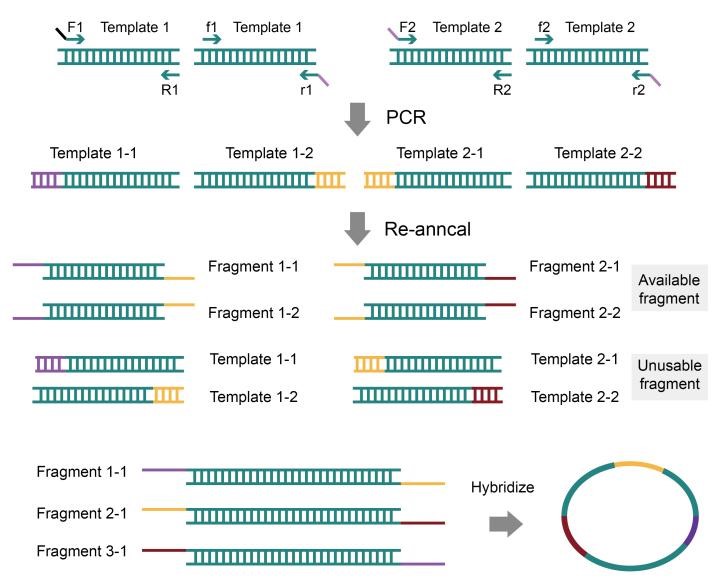
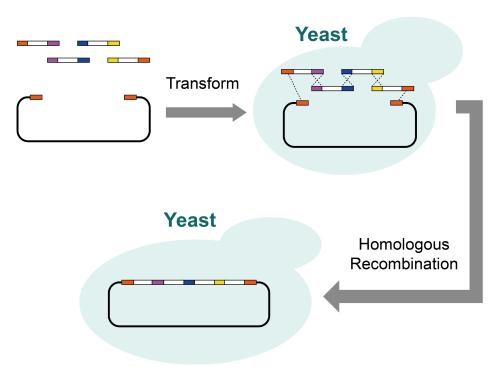
By enabling powerful genomics and biotechnology research, synthetic DNA assembly technology has revolutionized the field. From enzyme-dependent techniques to non-enzyme options and homologous recombination in yeast, a variety of modern methods are available—such as OE-PCR, BioBricks, Golden Gate Gibson with TPA (Twin Primer Non Enzymatic) at its core; all contributing to unprecedented advances in our understanding of genetic engineering.
Synbio Technologies is setting the standard as a leader in enabling technologies for synthetic biology. Our proprietary gene synthesis and assembly technology platform, combined with our groundbreaking bio-intelligent analysis tools such as Complexity Index (CI), NG Codon, AI-TAT and DNA Studio allow us to create tailored solutions that are smarter, faster, more precise – even those involving complex sequences of large scales including repetitive high GC or palindromic DNA (>10kb).
Case Study
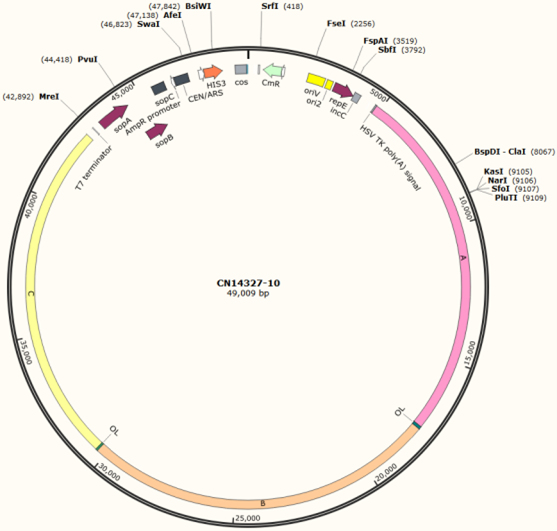
Synbio Technologies successfully achieved their customer’s goal of synthetically producing a 37,848bp target fragment and assembling it into the pSynoYac1-Cm (~ 10kb) — providing support for future biotechnology breakthroughs.
| Sequence Length | 37,848bp |
| Sequence Complexity | Extreme |
| GC Content | 42.3 | Normal |
| GC50nt Volatility | 60 | Complex |
| GC50nt Content | 70-10 | Complex |
| Base Repeatability | A10C5G6T17 | Complex |
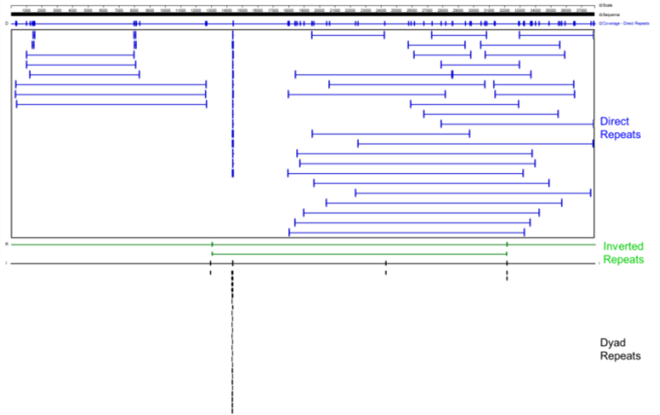
| Number of Long Repeats (20nt) | 108 | Extreme |
| Number of Short Repeats (9nt) | 29.2% | Normal |
| Palindromic Sequence Length | 16 | Normal |
| Reverse Repetition Length | 79 | Extreme |
Analysis of Difficulty
- Synthesis of this target fragment requires more than 400 small oligos and more than 80 times of overlapping extension PCR splicing assembly, which is a large workload with a low success rate.
- The complexity of the sequences is high, the GC content fluctuates, and there are up to 108 long, repeated sequences. (≥20 bp).
- The assembly of a 38kb fragment into a 10kb vector is very difficult and has a low success rate.
- The target fragment is highly homologous to the host cell genome, which is highly susceptible to fragment deletions as well as base mutations.
Solution
To combat the challenge of complex GC content, we fine-tuned our PCR reaction setup by manipulating polymerase amounts and ion concentrations. Then, to ensure reliable results, we further sharpened our parameters – like annealing temperature and extension time – for utmost efficiency in chain assembly during PCR amplification processes.
Drawing on CRISPR gene editing technology, we developed a modified brewer’s yeast strain (BY4741) that provided greater specificity and stability when it came to homologous recombination. Additionally, by refining the operating procedures for transformation as well as reaction system formulation with fragment deletion in mind, we achieved improved yield of large DNA fragments via our novel method surpassing existing kits commercially available today. Our optimized plasmid extraction technique proved successful allowing us to arrive at a robust yet stable end product ready for applications requiring high precision metrics and results from within the life sciences arena. We achieved success utilizing a two-round process of yeast in vivo homologous recombination assembly, culminating in the accurate construction of a 37,848bp target sequence.
Synbio Technologies provides you with solutions for the synthesis and assembly of long, complex DNA sequences. Your target sequences are delivered quickly and 100% accurate with a guaranteed cost advantage over other providers. Expertise. Accuracy. Savings. That’s what you get with Synbio Technologies.
Reference:
- Clarke L, Kitney R. Developing synthetic biology for industrial biotechnology applications. Biochem Soc Trans. 2020 Feb 28;48(1):113-122.
- Chang H, Wang C, Wang P, Zhou J, Li B. DNA assembly technologies: a review. Sheng Wu Gong Cheng Xue Bao. 2019 Dec 25;35(12):2215-2226.
- Clarke LJ, Kitney RI. Synthetic biology in the UK – An outline of plans and progress. Synth Syst Biotechnol. 2016 Oct 17;1(4):243-257.
- Chao R, Yuan Y, Zhao H. Recent advances in DNA assembly technologies. FEMS Yeast Res. 2015 Feb;15(1):1-9.
- Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349-61.
- Gibson DG, Benders GA, Axelrod KC, etc. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20404-9.