As synthetic biology experts, we can tell you that microbial genome editing is a cutting-edge technology that has revolutionized the field of biotechnology. By precisely modifying the DNA sequences of microorganisms, we can engineer strains that produce high-value products, such as biofuels, pharmaceuticals, and bioplastics. DNA synthesis has enabled us to perform highly efficient and precise genome editing, making it possible to create novel genetic circuits and metabolic pathways. With these tools and techniques, the possibilities for microbial genome editing are endless, and we are only just scratching the surface of what is possible.
Synbio Technologies is a leading provider of DNA sequencing, synthesis, and editing services, with a proven track record of success. By utilizing our synthesis and assembly platform, we can achieve efficient and reliable genome editing, based on the unique needs of every customer. With established genome editing systems in
E. coli and yeast cells, customers can be confident that their projects are in capable hands.
Contact us for about your next microbial genome editing project to save time, reduce costs, and ensure the highest quality results.
As a gene editing researcher, you demand the most precise and efficient tools available to ensure your research is successful. At Synbio Technologies, we provide a cutting-edge genome editing service that guarantees precision down to the base pair level, leaving no selection marker behind. Our genome editing tool is highly efficient, allowing you to edit any gene at any locus with the highest accuracy, and multiple sites can be mutated simultaneously. Our expert team utilizes the latest DNA technology to achieve these results, making us the leading choice for gene editing research. Trust Synbio Technologies to provide you with the most advanced genome editing technology available, and help take your research to the next level.
Service Specifications
| Service | E. coli Knock-out | E. coli Knock-in |
Yeast Knock-out |
Yeast Knock-in |
| Guide RNA Design | Optimizing guide RNA sequence for maximized efficiency and minimal off-target effects using design algorithms created at Synbio Technologies. | |||
| Materials Needed from Clients |
|
|||
| Deliverables | Engineered strain in glycerol stock | |||
| Quality Control |
|
|||
| Delivery Time | Starting from 4 weeks | |||
| Pricing | Starting from $3,500 | Starting from $4,500 | Starting from $4,900 | Starting from $5,900 |
*Pricing does not include the additional cost for gene synthesis, if required.
Case Study: Knocking fluorescent protein genes into E. coli
We have successfully knocked the 906bp red fluorescent protein coding gene into the genome of E.coli with a knock-in success rate as high as 96.6%~100%. This achievement demonstrates the power and precision of our gene editing technology, and we are excited to offer these capabilities to our clients. Trust Synbio Technologies to provide you with cutting-edge gene editing technology and expert consultation to ensure the success of your research.
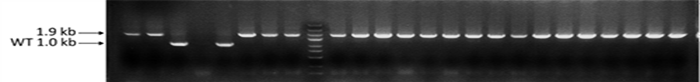
Fig. 1 PCR identification electrophoresis of clones after red fluorescent protein gene knock-in
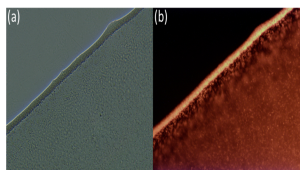
Fig. 2 Images of cells of E. coli mutant strain under fluorescence microscopy. (a) Picture of cells under white light, (b) Picture of cells after excitation by a 594 nm light source.
Case Study: Yeast Genome Editing
The ADE1 gene regulates the expression of adenine in yeast, and the engineered mutants displayed an accumulation of red pigment, which is a phenotype associated with a lack of adenine. To confirm the success of the gene modification, we conducted DNA sequencing, which revealed the absence of 18 base pairs, indicating a successful genetic modification (see fig. 2). These results showcase the accuracy and effectiveness of our gene editing technology and highlight the potential for this technique to advance research in biotechnology and other related fields.
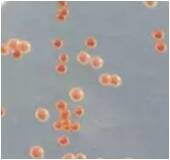
Fig.1. Programmed pink yeast colonies

Fig.2 DNA sequencing

